| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 186,364 – 186,463 |
| Length | 99 |
| Max. P | 0.900078 |

| Location | 186,364 – 186,463 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.07 |
| Mean single sequence MFE | -31.05 |
| Consensus MFE | -16.68 |
| Energy contribution | -17.60 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
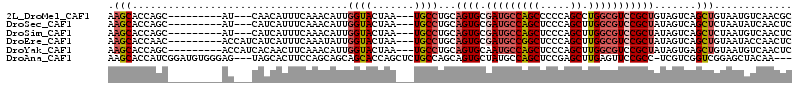
>2L_DroMel_CAF1 186364 99 + 22407834 AAGCACCAGC---------AU---CAACAUUUCAAACAUUGGUACUAA---UGCCUGCAGUGCGAUGCCAGCCCCCAGCCUGGCGUCCGCUGUAGUCAGCUGUAAUGUCAACGC ..(((.((((---------..---............(((((....)))---)).((((((((.((((((((........))))))))))))))))...))))...)))...... ( -32.80) >DroSec_CAF1 2607 99 + 1 AAGCACCAGC---------AU---CAUCAUUUCAAACAUUGGUACUAA---UGCCUGCAGUGCGAUGCCAGCUCCCAGCUUGGCGUCCGCUAUAGUCAGCUCUAAUAUCAACUC ..((....))---------..---..............((((((.((.---.(((((.((((.(((((((((.....)).))))))))))).)))...))..)).))))))... ( -26.20) >DroSim_CAF1 2599 99 + 1 AAGCACCAGC---------AU---CAUCAUUUCAAACAUUGGUACUAA---UGCCUGCAGUGCGAUGCCAGCUCCCAGCUUGGCGUCCGCUAUAGUCAGCUCUAAUGUCAACUC ..((....))---------..---...........(((((((......---((.(((.((((.(((((((((.....)).))))))))))).))).))...)))))))...... ( -28.10) >DroEre_CAF1 2623 102 + 1 AAGCACCAAC---------ACCAUCAUCAUUUCAAAUAUUGGUACUAA---UGCCUGCAGUGCGAUGCCGGCUCCCAGCUUGGCGUCCGCUAUAGUCAGCUGUAAUACCAACUC ..........---------...................((((((.((.---((.(((.((((.(((((((((.....)).))))))))))).))).))....)).))))))... ( -26.90) >DroYak_CAF1 2619 102 + 1 AAGCACCAGC---------ACCAUCACAACUUCAAACAUUGGUACUAA---UGCCUGCAGUGCAAUGCCAGCUCCCAGCUUGGCGUCCGCUAUAGUGAGCUGUAAUGUCAACUC ..(((.((((---------....((((.(((.((..(((((....)))---))..)).)))((.((((((((.....)).))))))..))....))))))))...)))...... ( -24.80) >DroAna_CAF1 1722 107 + 1 AAGCACCAUCGGAUGUGGGAG---UAGCACUUCCAGCAGCAGCACCAGCUCUGCCAGCAGUGCUAUGCCAGCUCCGAGCUUGAGUUCCGCC-UCGUCGGUCGGAGCUACAA--- ((((....(((((.((.((.(---(((((((.(..((((.(((....)))))))..).)))))))).)).))))))))))).(((((((((-.....)).)))))))....--- ( -47.50) >consensus AAGCACCAGC_________AU___CAUCAUUUCAAACAUUGGUACUAA___UGCCUGCAGUGCGAUGCCAGCUCCCAGCUUGGCGUCCGCUAUAGUCAGCUGUAAUGUCAACUC .(((....................................((((.......))))...((((.(((((((((.....)).))))))))))).......)))............. (-16.68 = -17.60 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:24:01 2006