| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,661,392 – 12,661,488 |
| Length | 96 |
| Max. P | 0.926765 |

| Location | 12,661,392 – 12,661,488 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 65.17 |
| Mean single sequence MFE | -21.39 |
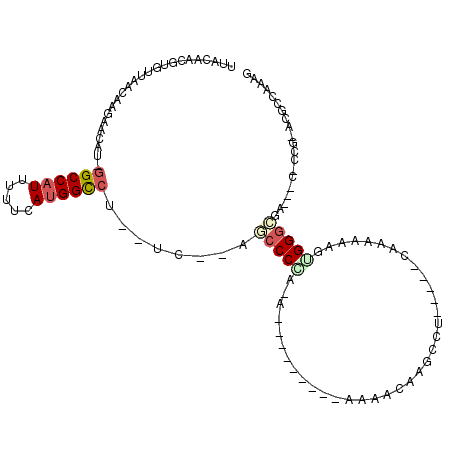
| Consensus MFE | -7.86 |
| Energy contribution | -8.39 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.37 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926765 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
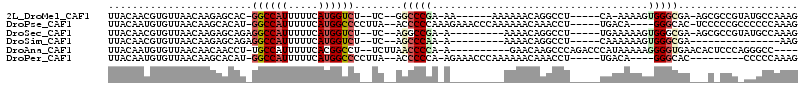
>2L_DroMel_CAF1 12661392 96 + 22407834 UUACAACGUGUUAACAAGAGCAC-GGCCAUUUUUCAUGGUCU--UC--GGCCCGA-AA------AAAAAACAGGCCU-----CA-AAAAGUGGGCGA-AGCGCCGUAUGCCAAAG .......(((((......)))))-((((((((((...(((((--((--(...)))-..------.......))))).-----..-)))))))((((.-..))))....))).... ( -23.70) >DroPse_CAF1 51943 102 + 1 UUACAAUGUGUUAACAAGCACAU-GGCCAUUUUUCAUGGCCCCUUA--ACCCCCAAAGAAACCCAAAAAACAAACCU-----UGACA----GGGCAC-UCCCCCGCCCCCCAAAG .....(((((((....)))))))-((((((.....)))))).....--...........................((-----(....----((((..-......))))....))) ( -21.80) >DroSec_CAF1 41955 95 + 1 UUACAACGUGUUAACAAGAGCAGAGGCCAUUUUUCAUGGUCU--UC--AGGCCGA-A---------AAAACAGGCCU-----UGAAAAAGUGGGCGA-AGCGCCGUAUGCCAAAG .................(.(((((((((.((((((..((((.--..--.))))))-)---------)))...)))))-----).........((((.-..))))...)))).... ( -25.10) >DroSim_CAF1 43539 81 + 1 UUACAACGUGUUAACAAGAGCAGAGGCCAUUUUUCAUGGUCU--UC--AGCCCAA-A---------AAAACAGGCCU-----CAAAAAAGUGGGCGA---------------AAG ......((((((......))).((((((..((((..(((.(.--..--.).))).-.---------))))..)))))-----)..........))).---------------... ( -18.10) >DroAna_CAF1 35119 97 + 1 UUACAAUGUGUUAACAACAACCU-UGCCAUUUUUCACGGCCU--UCUUAACCCCA-A----------GAACAAGCCCAGACCCAUAAAAAGGGGUGAACACUCCCAGGGCC---- ......(((....)))....(((-((......((((((((.(--((((......)-)----------)))...)))....(((.......))))))))......)))))..---- ( -20.60) >DroPer_CAF1 51572 93 + 1 UUACAAUGUGUUAACAAGCACAU-GGCCAUUUUUCAUGGCCCCUUA--ACCCCCA-AGAAACCCAAAAAACAAACCU-----UGACA----GGGCAC---------CCCCCAAAG .....(((((((....)))))))-((((((.....)))))).....--.....((-((.................))-----))...----(((...---------..))).... ( -19.03) >consensus UUACAACGUGUUAACAAGAACAU_GGCCAUUUUUCAUGGCCU__UC__AGCCCCA_A_________AAAACAAGCCU_____CAAAAAAGUGGGCGA___C_CCG_ACGCCAAAG ........................((((((.....))))))........(((((....................................))))).................... ( -7.86 = -8.39 + 0.53)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:57 2006