
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,655,894 – 12,656,025 |
| Length | 131 |
| Max. P | 0.913350 |

| Location | 12,655,894 – 12,655,987 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 82.17 |
| Mean single sequence MFE | -25.72 |
| Consensus MFE | -15.12 |
| Energy contribution | -15.93 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.08 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12655894 93 - 22407834 AAUAUAUCCAACAA-GCG------CGCUCUCUCCCGCACAAACACACUCACCCAAAGUGGGCAGCCGGCAGCCAAUCGGCAUGGAGCUUUGGCGCUCUCU .............(-(((------((((((..(((((...................)))))..(((((.......)))))..)))))....))))).... ( -24.81) >DroSec_CAF1 36900 93 - 1 AAUACAUCCAACAA-GCG------CGCUCUCUCCCACACACCCACACACACCCAAAGUGGGCAGCCGGCAGCCAAUCGGCAUGGAGCUUUGGCGCUCUCU .............(-(.(------((((..((((......(((((...........)))))..(((((.......)))))..))))....))))).)).. ( -27.00) >DroSim_CAF1 37608 93 - 1 AAUAUAUCCAACAA-GCG------CGCUCUCUCCCACACACCCACACUCAACCAAAGUGGGCAGCCGGCAGCCAAUCGGCAUGGAGCUUUGGCGCUCUCU .............(-(.(------((((..((((......(((((...........)))))..(((((.......)))))..))))....))))).)).. ( -27.00) >DroEre_CAF1 37198 93 - 1 AAUAUAUCCAACGA-GCG------CGCUCUCUCCCACACACCCACACUCACUCAAAGUGGGCAGCCGGCAGCCAAUCGGCUUGAAGCUUUGGCGCUCUCU ............((-(((------((((..(((((((...................))))).))..)))((((....))))..........))))))... ( -28.41) >DroYak_CAF1 38422 93 - 1 AAUAUAUCCAACGA-GCG------CGCUCUCUCCCACACACCUACACUUACUCAAAGUGGGCAGCAGGCAGCCAAUGGGCUUGGAGCUUUGGCGCUCUCU ............((-(((------((((((..((((........(((((.....)))))(((........)))..))))...)))))....))))))... ( -31.30) >DroAna_CAF1 30378 94 - 1 AAUGUUCACAUUCACUCUCUUUCUCUCUCUCUCUUCCACUUACACACUGA--AAAGGUGGG----CGACAGCCAAUCGUUUUGGAGCUUUGACGCUCUCA ..........................................(((.((..--..)))))((----(....))).........(((((......))))).. ( -15.80) >consensus AAUAUAUCCAACAA_GCG______CGCUCUCUCCCACACACCCACACUCACCCAAAGUGGGCAGCCGGCAGCCAAUCGGCAUGGAGCUUUGGCGCUCUCU ...............((........(((..(((((((...................))))).))..))).(((((..(((.....))))))))))..... (-15.12 = -15.93 + 0.81)

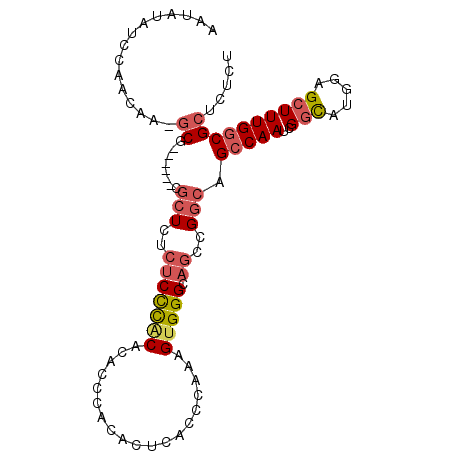

| Location | 12,655,929 – 12,656,025 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 77.71 |
| Mean single sequence MFE | -28.82 |
| Consensus MFE | -15.77 |
| Energy contribution | -17.33 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12655929 96 - 22407834 A--GAGAAAGUGCGGAGGGAGAGGUAGAAAAUCUUGGGGAAAUAUAUCCAACAA-GCG------CGCUCUCUCCCGCACAAACACACUCACCCAAAGUGGGCAGC .--......((((((.(((((((((.......((((.(((......)))..)))-).)------).))))))))))))).......(((((.....))))).... ( -31.60) >DroSec_CAF1 36935 98 - 1 AGAGAGAAAGUGCGGAGGGAGAGGUUGAAAAUCUUGGGGAAAUACAUCCAACAA-GCG------CGCUCUCUCCCACACACCCACACACACCCAAAGUGGGCAGC .........(((.((((((((.(.(((......(((((........))))))))-.).------..)))))))))))...(((((...........))))).... ( -30.00) >DroSim_CAF1 37643 96 - 1 A--GAGAAAGUGCGGAGGGAGAGGUUGAAAAUCUUGGGGAAAUAUAUCCAACAA-GCG------CGCUCUCUCCCACACACCCACACUCAACCAAAGUGGGCAGC .--......(((.((((((((.(.(((......(((((........))))))))-.).------..)))))))))))...(((((...........))))).... ( -30.00) >DroEre_CAF1 37233 96 - 1 A--GAGAAAGUGCGGAGGGAAAGGCAGAAAAUCUUGGGGAAAUAUAUCCAACGA-GCG------CGCUCUCUCCCACACACCCACACUCACUCAAAGUGGGCAGC .--......(((.(((((((...((.(....(((((((........))))).))-.).------.))))))))))))...(((((...........))))).... ( -27.30) >DroYak_CAF1 38457 96 - 1 A--GAGAAAGUGCGGAGGGAGAGGCAGAAAAUCUUGGGGAAAUAUAUCCAACGA-GCG------CGCUCUCUCCCACACACCUACACUUACUCAAAGUGGGCAGC .--......(((.((.(((((((((.(....(((((((........))))).))-.))------).))))))))).))).(((((...........))))).... ( -29.90) >DroAna_CAF1 30413 87 - 1 G--GAGGA-----GAAUAAAAAUCUUGAAA-----GGGGAAAUGUUCACAUUCACUCUCUUUCUCUCUCUCUCUUCCACUUACACACUGA--AAAGGUGGG---- (--(((((-----((...........((((-----((((.((((....))))...))))))))......)))))))).....(((.((..--..)))))..---- ( -24.13) >consensus A__GAGAAAGUGCGGAGGGAGAGGUAGAAAAUCUUGGGGAAAUAUAUCCAACAA_GCG______CGCUCUCUCCCACACACCCACACUCACCCAAAGUGGGCAGC .........(((.((((((((............(((.(((......)))..)))............)))))))))))...(((((...........))))).... (-15.77 = -17.33 + 1.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:56 2006