
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,650,605 – 12,650,750 |
| Length | 145 |
| Max. P | 0.958094 |

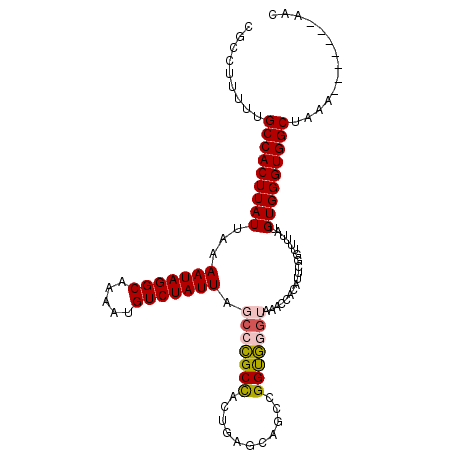
| Location | 12,650,605 – 12,650,713 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 80.81 |
| Mean single sequence MFE | -34.05 |
| Consensus MFE | -25.90 |
| Energy contribution | -25.85 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12650605 108 + 22407834 CGCCUUUUUGCCACUUAUUAAAAUAGGCAAAAUGUCUAUUAGGCCGCCAGUGAGCAGCCGGUGGGUAAACCACAUUCGUUUUUAUGUGGGUGGCUAAAAAAAAAAAAC ....((((((((((.......(((((((.....)))))))...(((((.((.....)).))))).....((((((........))))))))))).)))))........ ( -32.70) >DroPse_CAF1 38588 94 + 1 CGCCUUUUUGCCACUUAUUAAAAUAGGCAAAAUGUCUAUUAGCCUGCUUCUUGGUAGCUGGCAUGGAAAA-A---AAGGUACCAUGUGGGUGGCUGG----------C .(((.....(((((((((...(((((((.....))))))).(((.(((.......))).)))((((....-.---......))))))))))))).))----------) ( -31.40) >DroSec_CAF1 31716 107 + 1 CGCCUUUUUGCCACUUAUUAAAAUAGGCAAAAUGUCUAUUAGCCCGCCAGUGAGCAGCCGGUGGGUAAACCUCAUUCGUUUUUAUGUGGGUGGCUAGAAA-AAAAAAC ....((((((((((((((...(((((((.....))))))).(((((((.((.....)).)))))))...................))))))))).)))))-....... ( -33.80) >DroEre_CAF1 31767 101 + 1 CGCCUUUUUGCCACUUAUUAAAAUAGGCAAAAUGUCUAUUAGCCCGCCAUUGAGCAGCCGGUGGGUAAACCACAUUCGGUUUUAUGUGGGUGGCUGAA-------AGC ...(((((.(((((.......(((((((.....))))))).(((((((...........)))))))...((((((........))))))))))).)))-------)). ( -37.60) >DroYak_CAF1 32982 103 + 1 CGCCUUUUUGCCACUUAUUAAAAUAGGCAAAAUGUCUAUUAGCCCGCCGUUGAGCAGCCGGUGGGUAAACCACAUUCGGUUUUAUGUGGGUGGCUAAA-----AAAAC ....((((((((((.......(((((((.....))))))).((((((((((....)).))))))))...((((((........))))))))))).)))-----))... ( -37.50) >DroPer_CAF1 38266 95 + 1 CGCCUUUUUGCCACUUAUUAAAAUAGGCAAAAUGUCUAUUAGCCUGCUUCUUGGUAGCUGGCAUGGAAAAAA---AAGGUACCAUGUGGGUGGCUGG----------C .(((.....(((((((((...(((((((.....))))))).(((.(((.......))).)))((((......---......))))))))))))).))----------) ( -31.30) >consensus CGCCUUUUUGCCACUUAUUAAAAUAGGCAAAAUGUCUAUUAGCCCGCCACUGAGCAGCCGGUGGGUAAACCACAUUCGGUUUUAUGUGGGUGGCUAAA_______AAC .........(((((((((...(((((((.....))))))).(((((((...........)))))))...................))))))))).............. (-25.90 = -25.85 + -0.05)

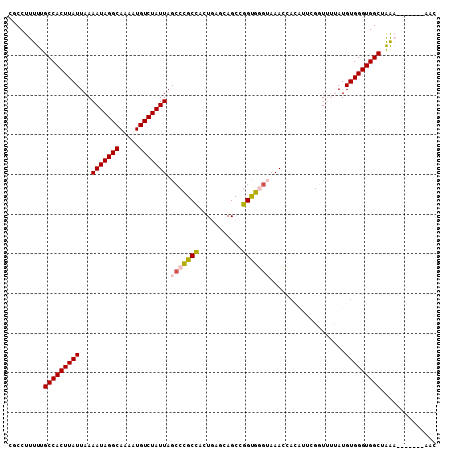
| Location | 12,650,605 – 12,650,713 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 80.81 |
| Mean single sequence MFE | -28.76 |
| Consensus MFE | -19.41 |
| Energy contribution | -20.02 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12650605 108 - 22407834 GUUUUUUUUUUUUAGCCACCCACAUAAAAACGAAUGUGGUUUACCCACCGGCUGCUCACUGGCGGCCUAAUAGACAUUUUGCCUAUUUUAAUAAGUGGCAAAAAGGCG ..............(((..((((((........))))))..........(((((((....))))))).........(((((((.((((....))))))))))).))). ( -32.20) >DroPse_CAF1 38588 94 - 1 G----------CCAGCCACCCACAUGGUACCUU---U-UUUUCCAUGCCAGCUACCAAGAAGCAGGCUAAUAGACAUUUUGCCUAUUUUAAUAAGUGGCAAAAAGGCG (----------((((((.....(((((......---.-....)))))...(((.......))).))))........(((((((.((((....))))))))))).))). ( -27.20) >DroSec_CAF1 31716 107 - 1 GUUUUUU-UUUCUAGCCACCCACAUAAAAACGAAUGAGGUUUACCCACCGGCUGCUCACUGGCGGGCUAAUAGACAUUUUGCCUAUUUUAAUAAGUGGCAAAAAGGCG .......-......(((.....................((((((((.((((.......)))).)))....))))).(((((((.((((....))))))))))).))). ( -26.40) >DroEre_CAF1 31767 101 - 1 GCU-------UUCAGCCACCCACAUAAAACCGAAUGUGGUUUACCCACCGGCUGCUCAAUGGCGGGCUAAUAGACAUUUUGCCUAUUUUAAUAAGUGGCAAAAAGGCG (((-------(..((((..(((.......(((...((((.....)))))))........)))..))))........(((((((.((((....))))))))))))))). ( -28.26) >DroYak_CAF1 32982 103 - 1 GUUUU-----UUUAGCCACCCACAUAAAACCGAAUGUGGUUUACCCACCGGCUGCUCAACGGCGGGCUAAUAGACAUUUUGCCUAUUUUAAUAAGUGGCAAAAAGGCG (((((-----(((.(((((((((((........))))))....(((.(((.........))).)))..(((((.(.....).))))).......))))))))))))). ( -31.40) >DroPer_CAF1 38266 95 - 1 G----------CCAGCCACCCACAUGGUACCUU---UUUUUUCCAUGCCAGCUACCAAGAAGCAGGCUAAUAGACAUUUUGCCUAUUUUAAUAAGUGGCAAAAAGGCG (----------((((((.....(((((......---......)))))...(((.......))).))))........(((((((.((((....))))))))))).))). ( -27.10) >consensus GUU_______UCCAGCCACCCACAUAAAACCGAAUGUGGUUUACCCACCGGCUGCUCAAUGGCGGGCUAAUAGACAUUUUGCCUAUUUUAAUAAGUGGCAAAAAGGCG ..............(((.....................(((((....((.(((.......))).))....))))).(((((((.((((....))))))))))).))). (-19.41 = -20.02 + 0.61)


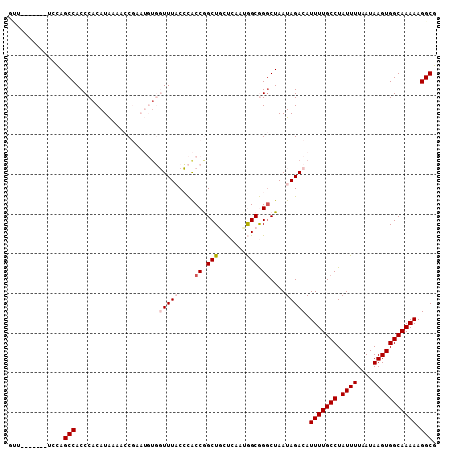
| Location | 12,650,645 – 12,650,750 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 74.74 |
| Mean single sequence MFE | -31.94 |
| Consensus MFE | -15.95 |
| Energy contribution | -16.01 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12650645 105 + 22407834 AGGCCGCCAGUGAGCAGCCGGUGGGUAAACCACAUUCGUUUUUAUGUGGGUGGCUAAAAAAAAAAAACACCUGGAAACGGUGAGG-GAAUGGAAUGGAAAACCCAC .(((((((.(((((.(((..((((.....))))....))))))))...)))))))............((((.(....))))).((-(..............))).. ( -33.04) >DroSec_CAF1 31756 104 + 1 AGCCCGCCAGUGAGCAGCCGGUGGGUAAACCUCAUUCGUUUUUAUGUGGGUGGCUAGAAA-AAAAAACACCUGGAAACGGUGAGG-GAAUGGAAUGGAAAACCCAC .(((((((.((.....)).)))))))...((((((.(((((((....(((((........-......))))))))))))))))))-........(((.....))). ( -35.34) >DroSim_CAF1 32414 104 + 1 AGCCCGCCAGUGAGCAGCCGGUGGGUAAACCACAUUCGUUUUUAUGUGGGUGGCUAGAAA-AAAAAACACCUGGAAACGGUGAGG-GAAUGGCAUGGAAAACCCAC .(((((((.((.....)).)))))))...((((((........))))))...((((....-......((((.(....)))))...-...)))).(((.....))). ( -33.09) >DroEre_CAF1 31807 94 + 1 AGCCCGCCAUUGAGCAGCCGGUGGGUAAACCACAUUCGGUUUUAUGUGGGUGGCUGAA-------AGCACCUGGAAUCGUAAGGG-GAAUGG----AGAAACCCAG .(((((((...........)))))))(((((......)))))....(((((.((....-------.)).((.....((....)).-....))----....))))). ( -27.00) >DroYak_CAF1 33022 95 + 1 AGCCCGCCGUUGAGCAGCCGGUGGGUAAACCACAUUCGGUUUUAUGUGGGUGGCUAAA-----AAAACACCUGGAAACGCUGAGG-GAAUG-----AAAAACCGAC .((((((((((....)).)))))))).........((((((((....(((((......-----....)))))(....).......-.....-----.)))))))). ( -30.70) >DroPer_CAF1 38306 92 + 1 AGCCUGCUUCUUGGUAGCUGGCAUGGAAAAAA---AAGGUACCAUGUGGGUGGCUGG----------CAACUGGAGGUGGCGCGGUGGGUGCGGUGG-GAGCCCAG .(((..(((((.(((.(((.((((((......---......)))))).))).)))(.----------...).)))))..).))..(((((.(.....-).))))). ( -32.50) >consensus AGCCCGCCAGUGAGCAGCCGGUGGGUAAACCACAUUCGGUUUUAUGUGGGUGGCUAAA_____AAAACACCUGGAAACGGUGAGG_GAAUGG_AUGGAAAACCCAC .(((((((...........)))))))..........(((((((....(((((...............))))))))))))........................... (-15.95 = -16.01 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:53 2006