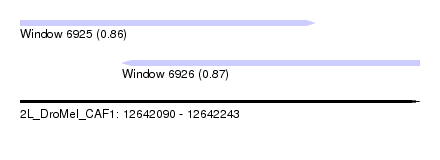
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,642,090 – 12,642,243 |
| Length | 153 |
| Max. P | 0.867171 |

| Location | 12,642,090 – 12,642,203 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 93.51 |
| Mean single sequence MFE | -24.42 |
| Consensus MFE | -19.73 |
| Energy contribution | -20.85 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12642090 113 + 22407834 UGGGGUUUUCAUAUUUUUCGAAUAUUUACCAUCGAUUAUGUGGUUAAUUUUCCAUUGAUAUUCAGGCACAUAAAGUUUCCGACCUGUUCAUAAAUUUUUAUGAACAUUUUAAA ..(((((...(((((..(.(((.(((.(((((.......))))).))).))).)..)))))...((.((.....))..)))))))((((((((....))))))))........ ( -20.30) >DroSec_CAF1 23172 113 + 1 UGGGGUUUUCAUAUUUUUGGAAUAUUUACCAUCGAUUAUAUGGUUAAUUUUCCAUUGAUAUUCAGGCUCAUAAAGUUUCGGACCUGUUCAUAAAUGUUUAUGAAAAUUUUUAA .((((((((((((....(((((.(((.(((((.......))))).))).))))).(((....((((..(..........)..)))).)))........))))))))))))... ( -24.70) >DroSim_CAF1 23892 113 + 1 UCGGGUUUUCAUAUUUUUGGAAUAUUUACCAUCGAUUAUGUGGUUAAUUUUCCAUUGAUAUUCAGGCUCAUAAAGUUUCCGACCUGUUCAUAAAUGUUUAUGAACAUUUUUAA ..((((((..(((((..(((((.(((.(((((.......))))).))).)))))..)))))..))))))...............(((((((((....)))))))))....... ( -28.80) >DroYak_CAF1 23945 112 + 1 UGGGGUUUUCAUAUU-UUGGAAUAUUUACCGUCGAUUAUGUGCUUAAUUUUCCAUUGAUAUUCCGGGUCAUAAAAUUUCCGACCUGUUCAUAAAUUUUUAUGAACAUUUUAAA .(..(((((.((..(-((((((((((....(..(((((......)))))..)....)))))))))))..)))))))..).....(((((((((....)))))))))....... ( -23.90) >consensus UGGGGUUUUCAUAUUUUUGGAAUAUUUACCAUCGAUUAUGUGGUUAAUUUUCCAUUGAUAUUCAGGCUCAUAAAGUUUCCGACCUGUUCAUAAAUGUUUAUGAACAUUUUAAA ..((((((..(((((..(((((.(((.(((((.......))))).))).)))))..)))))..))))))...............(((((((((....)))))))))....... (-19.73 = -20.85 + 1.12)

| Location | 12,642,129 – 12,642,243 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 86.01 |
| Mean single sequence MFE | -19.62 |
| Consensus MFE | -14.15 |
| Energy contribution | -15.28 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.867171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12642129 114 - 22407834 UCGCAAGGAAACGAGACAGCUUCAUAAACAUAAAUAAGUUUUUAAAAUGUUCAUAAAAAUUUAUGAACAGGUCGGAAACUUUAUGUGCCUGAAUAUCAAUGGAAAAUUAACCAC ......(....)........((((...((((((...((((((..(..(((((((((....)))))))))..)..))))))))))))...))))......(((........))). ( -24.90) >DroSec_CAF1 23211 110 - 1 ----AAGAAGACAAGAUAGCCUUAUAAACAUAAAUAAGUUUUAAAAAUUUUCAUAAACAUUUAUGAACAGGUCCGAAACUUUAUGAGCCUGAAUAUCAAUGGAAAAUUAACCAU ----..........((((..(((((........)))))...........(((((((....)))))))((((..((........))..))))..)))).((((........)))) ( -15.60) >DroSim_CAF1 23931 110 - 1 ----AAGAAGACAAGAUAGCCUUAUAAACAUAAAUAAGUUUUAAAAAUGUUCAUAAACAUUUAUGAACAGGUCGGAAACUUUAUGAGCCUGAAUAUCAAUGGAAAAUUAACCAC ----............(((.(((((((.........((((((..(..(((((((((....)))))))))..)..))))))))))))).)))........(((........))). ( -19.90) >DroYak_CAF1 23983 104 - 1 ----AAGGCGUAAAGAU------AUAAAGAUAUAUAAGUUUUUAAAAUGUUCAUAAAAAUUUAUGAACAGGUCGGAAAUUUUAUGACCCGGAAUAUCAAUGGAAAAUUAAGCAC ----..((((((((.((------(((....)))))..(((((..(..(((((((((....)))))))))..)..))))))))))).)).......................... ( -18.10) >consensus ____AAGAAGACAAGAUAGCCUUAUAAACAUAAAUAAGUUUUAAAAAUGUUCAUAAAAAUUUAUGAACAGGUCGGAAACUUUAUGAGCCUGAAUAUCAAUGGAAAAUUAACCAC ................(((.(((((((.........((((((..(..(((((((((....)))))))))..)..))))))))))))).)))....................... (-14.15 = -15.28 + 1.13)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:51 2006