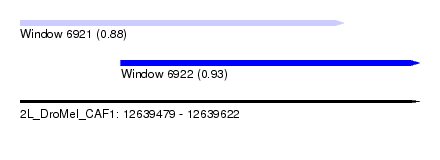
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,639,479 – 12,639,622 |
| Length | 143 |
| Max. P | 0.925374 |

| Location | 12,639,479 – 12,639,595 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.60 |
| Mean single sequence MFE | -38.17 |
| Consensus MFE | -30.33 |
| Energy contribution | -30.50 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
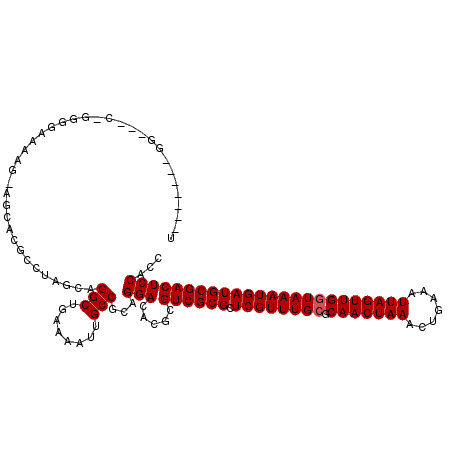
>2L_DroMel_CAF1 12639479 116 + 22407834 UGA----GGUGAUCGGGGGAAAUGAAGCACGCCUAGCAGGCUGAAAAUUGCCGCAGGACACGCGUCGCGCUCGUUUGCGCAACUAAACUGAAAUUAGUUGGUAAAUGAUGCGACUCCACC ...----.......((.(((......((...(((.((.(((........))))))))....))((((((.((((((((.(((((((.......)))))))))))))))))))))))).)) ( -42.00) >DroPse_CAF1 24287 91 + 1 ---------------------------CAGGCUUUG--GGCUGAAAACUGCCGCAGGACACGCGUCGCGCUCGUUUGCGCAACUAAACUGAAAUUAGUUGGUAAAUGAUGCGACUCCACC ---------------------------..((.((((--(((........))).))))......((((((.((((((((.(((((((.......))))))))))))))))))))).))... ( -32.10) >DroEre_CAF1 20666 114 + 1 UA------GGCGGCCGGGGAAAAGGAGCACGCCUAGCAGGCUGAAAAUUGCCGCAGGACACGCGUCGCGCUCGUUUGCGCAACUAAGCUGAAAUUAGUUGGUAAAUGAUGCGACUCCACC ((------((((.((........))....))))))((.(((........))))).(((.....((((((.((((((((.(((((((.......))))))))))))))))))))))))... ( -45.80) >DroYak_CAF1 21225 114 + 1 AA------GGGGGCCGGGGAAAAGGAGCACGCCUAGCAGGCUGAAAAUUGCCGCAGGACACGCGUCGCGCUCGUUUGCGCAACUAAACUGAAAUUAGUUGGUAAAUGAUGCGACUCCACC ..------.......(((((......((...(((.((.(((........))))))))....))((((((.((((((((.(((((((.......)))))))))))))))))))))))).)) ( -41.90) >DroAna_CAF1 15122 119 + 1 UGGAGUUGGGAAAA-AGGGGAAAGCAGUACGCCUAGCAGGCUGAAAAUUGCCACAGGACACGCGUCGCGCUCGUUUGGGCAACUAAACUGAAAUUAGUUGGUAAAUGAUGCGACUCCACC ((((((((((....-..(......)......)))))).(((........)))...........((((((.((((((.(.(((((((.......))))))).))))))))))))))))).. ( -35.10) >DroPer_CAF1 24242 91 + 1 ---------------------------CAGGCUUUG--GGCUGAAAACUGCCGCAGGACACGCGUCGCGCUCGUUUGCGCAACUAAACUGAAAUUAGUUGGUAAAUGAUGCGACUCCACC ---------------------------..((.((((--(((........))).))))......((((((.((((((((.(((((((.......))))))))))))))))))))).))... ( -32.10) >consensus U_______GG___C_GGGGAAAAG_AGCACGCCUAGCAGGCUGAAAAUUGCCGCAGGACACGCGUCGCGCUCGUUUGCGCAACUAAACUGAAAUUAGUUGGUAAAUGAUGCGACUCCACC ......................................(((........)))...(((.....((((((.((((((((.(((((((.......))))))))))))))))))))))))... (-30.33 = -30.50 + 0.17)

| Location | 12,639,515 – 12,639,622 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.56 |
| Mean single sequence MFE | -29.65 |
| Consensus MFE | -25.95 |
| Energy contribution | -26.28 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
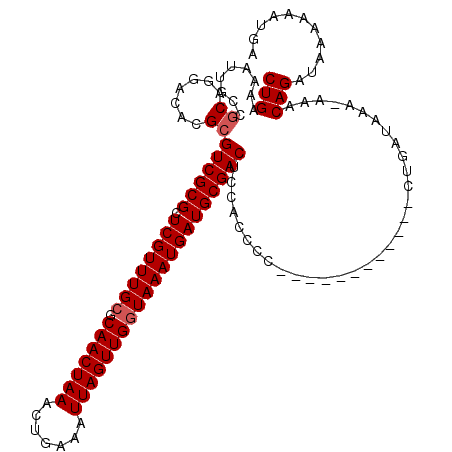
>2L_DroMel_CAF1 12639515 107 + 22407834 CUGAAAAUUGCCGCAGGACACGCGUCGCGCUCGUUUGCGCAACUAAACUGAAAUUAGUUGGUAAAUGAUGCGACUCCACCCU------------CUGAUAAA-AAACAGAUAAAAAAUGA (((....(((.((.(((....(.((((((.((((((((.(((((((.......))))))))))))))))))))).)...)))------------.)).))).-...)))........... ( -28.80) >DroPse_CAF1 24298 119 + 1 CUGAAAACUGCCGCAGGACACGCGUCGCGCUCGUUUGCGCAACUAAACUGAAAUUAGUUGGUAAAUGAUGCGACUCCACCCCCCGUCCAGAGCCAUGAUAAA-ACACAGAUAAAAAAUGA (((....(((.((..((....(.((((((.((((((((.(((((((.......))))))))))))))))))))).)...))..))..))).....((.....-.)))))........... ( -29.60) >DroEre_CAF1 20700 107 + 1 CUGAAAAUUGCCGCAGGACACGCGUCGCGCUCGUUUGCGCAACUAAGCUGAAAUUAGUUGGUAAAUGAUGCGACUCCACCCU------------CUGAUAAA-AAACAGAUAAAAAAUGA (((....(((.((.(((....(.((((((.((((((((.(((((((.......))))))))))))))))))))).)...)))------------.)).))).-...)))........... ( -29.60) >DroYak_CAF1 21259 108 + 1 CUGAAAAUUGCCGCAGGACACGCGUCGCGCUCGUUUGCGCAACUAAACUGAAAUUAGUUGGUAAAUGAUGCGACUCCACCCU------------CUGAUAAAAAAACAGAUAAAAAAUGA (((....(((.((.(((....(.((((((.((((((((.(((((((.......))))))))))))))))))))).)...)))------------.)).))).....)))........... ( -28.20) >DroAna_CAF1 15161 117 + 1 CUGAAAAUUGCCACAGGACACGCGUCGCGCUCGUUUGGGCAACUAAACUGAAAUUAGUUGGUAAAUGAUGCGACUCCACCCCC--UCCUGGGCGCUGAUAAA-AAACAGAUAAAAAAUGA .........(((.(((((...(.((((((.((((((.(.(((((((.......))))))).))))))))))))).).......--)))))))).(((.....-...)))........... ( -32.10) >DroPer_CAF1 24253 119 + 1 CUGAAAACUGCCGCAGGACACGCGUCGCGCUCGUUUGCGCAACUAAACUGAAAUUAGUUGGUAAAUGAUGCGACUCCACCCCCCGUCCAGAGCCAUGAUAAA-ACACAGAUAAAAAAUGA (((....(((.((..((....(.((((((.((((((((.(((((((.......))))))))))))))))))))).)...))..))..))).....((.....-.)))))........... ( -29.60) >consensus CUGAAAAUUGCCGCAGGACACGCGUCGCGCUCGUUUGCGCAACUAAACUGAAAUUAGUUGGUAAAUGAUGCGACUCCACCCC____________CUGAUAAA_AAACAGAUAAAAAAUGA (((.........((.......))((((((.((((((((.(((((((.......)))))))))))))))))))))................................)))........... (-25.95 = -26.28 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:47 2006