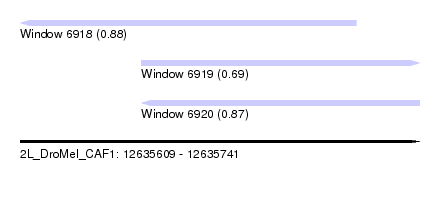
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,635,609 – 12,635,741 |
| Length | 132 |
| Max. P | 0.883586 |

| Location | 12,635,609 – 12,635,720 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.26 |
| Mean single sequence MFE | -39.02 |
| Consensus MFE | -26.49 |
| Energy contribution | -26.55 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12635609 111 - 22407834 CUCCCAGCCGCAACGCAAAUCUUUUGGGCUACUGUUGGGAGAC--------CCGGAGCUCUUGUCCCUGGCCCCAUCCAUCGGCUUUGUGGCCUUUGAAGUCGCUCGUUGCGGUAGCAA ......(((((((((.....((((.(((((((.((((((.(.(--------(.((..(....)..)).))))))).......))...)))))))..)))).....)))))))))..... ( -41.61) >DroPse_CAF1 18968 89 - 1 CUUUGUGCCGCAACGCAAAUCUUUUGGGCUACUGUUGGGAGAG--------UC----------------------UUCUUCGGCUUUUGUGCCUUUGAAGUCGAUUGUUGCGGUAGCAA ..((((((((((((.....(((((..(.(....))..)))))(--------.(----------------------(((...(((......)))...)))).)....))))))))).))) ( -30.90) >DroSim_CAF1 17035 111 - 1 CUCCCAGCCGCAACGCAAAUCUUUUGGGCUACUGUUGGGAGAC--------CCGGAGCUCUUGUCCCUGGCCCCAUCCAUCGGCUUUGGGGCCUUUGAAGUCGCUCGUUGCGGUAGCAA ......(((((((((.....((((.(((((.(((..((....)--------)))))))))........(((((((.((...))...)))))))...)))).....)))))))))..... ( -44.90) >DroEre_CAF1 16746 110 - 1 CUCCCAGCCGCAACGCAAAUCUUUUGGGCUACUGUUGGGAGAC--------CCGGACCUCAUGUCCCUGGCCCCAUCCAUCGGCUUUG-GGCCUUUGAAGUCGCUCGUUGCGGUAGCAA ......(((((((((..........((((((.....((....)--------).((((.....)))).)))))).......((((((..-(....)..))))))..)))))))))..... ( -40.30) >DroYak_CAF1 17275 118 - 1 CUCCCAGCCGCAACGCAAAUCUUUUGGGCUACUGUUGGGAGACCCGGAGACCCGGAGCUCUUGUCCCUGGCCCCAUCCAUCGGCUUUG-AGCCUUUGAAGUCGCUCGUUGCGGUAGCAA ......(((((((((..........((((((..(.((((((..((((....))))..)))))).)..)))))).......((((((..-(....)..))))))..)))))))))..... ( -45.50) >DroPer_CAF1 18995 89 - 1 CUUUGUGCCGCAACGCAAAUCUUUUGGGCUACUGUUGGGAGAG--------UC----------------------UUCUUCGGCUUUUGUGCCUUUGAAGUCGAUUGUUGCGGUAGCAA ..((((((((((((.....(((((..(.(....))..)))))(--------.(----------------------(((...(((......)))...)))).)....))))))))).))) ( -30.90) >consensus CUCCCAGCCGCAACGCAAAUCUUUUGGGCUACUGUUGGGAGAC________CCGGAGCUCUUGUCCCUGGCCCCAUCCAUCGGCUUUGGGGCCUUUGAAGUCGCUCGUUGCGGUAGCAA ......(((((((((.....((((.(((((...(((((..((...........(((.......))).........))..))))).....)))))..)))).....)))))))))..... (-26.49 = -26.55 + 0.06)



| Location | 12,635,649 – 12,635,741 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 86.78 |
| Mean single sequence MFE | -40.82 |
| Consensus MFE | -30.59 |
| Energy contribution | -30.95 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12635649 92 + 22407834 UGGAUGGGGCCAGGGACAAGAGCUCCGG---------GUCUCCCAACAGUAGCCCAAAAGAUUUGCGUUGCGGCUGGGAGCCAAA--GUCCCUGGCCAGAACC .((....((((((((((....(((((.(---------(((...((((.((((..(....)..)))))))).)))).)))))....--))))))))))....)) ( -43.10) >DroSec_CAF1 16666 91 + 1 UG-AUGGGGCCAGGGACAAGAGCUCCGG---------GUCUCCCAACAGUAGUCCAAAAGAUUUGCGUUGCGGCUGGGAGCCAAA--GUCCCUGGGCAGAACC ..-....(.((((((((....(((((.(---------(((...((((.((((((.....)).)))))))).)))).)))))....--)))))))).)...... ( -39.10) >DroSim_CAF1 17075 92 + 1 UGGAUGGGGCCAGGGACAAGAGCUCCGG---------GUCUCCCAACAGUAGCCCAAAAGAUUUGCGUUGCGGCUGGGAGCCAAA--GUCCCUGGCCAGAACC .((....((((((((((....(((((.(---------(((...((((.((((..(....)..)))))))).)))).)))))....--))))))))))....)) ( -43.10) >DroEre_CAF1 16785 92 + 1 UGGAUGGGGCCAGGGACAUGAGGUCCGG---------GUCUCCCAACAGUAGCCCAAAAGAUUUGCGUUGCGGCUGGGAGCCAAA--GUCCCUGGCCAGAACC .((....((((((((((.((.....))(---------(.((((((.(.(((((.(((.....))).))))).).))))))))...--))))))))))....)) ( -42.80) >DroYak_CAF1 17314 100 + 1 UGGAUGGGGCCAGGGACAAGAGCUCCGGG-UCUCCGGGUCUCCCAACAGUAGCCCAAAAGAUUUGCGUUGCGGCUGGGAGCCAAA--GUCCCUGGCCAGAGCC .((....((((((((((....(((((.((-((...(((...)))....(((((.(((.....))).))))))))).)))))....--))))))))))....)) ( -50.10) >DroAna_CAF1 11479 103 + 1 UGGCUGUGACCAGGGACAGUAGCACCAGCAGCCACAUACAUCCCAACAGUAGCCCAAAAGAUUUGCGUUGCGGCCCAAAGCCAAAGAGUCCCAGGCCAGAAAC (((((..((((.((((..((.((.......)).)).....))))....(((((.(((.....))).)))))(((.....)))...).)))...)))))..... ( -26.70) >consensus UGGAUGGGGCCAGGGACAAGAGCUCCGG_________GUCUCCCAACAGUAGCCCAAAAGAUUUGCGUUGCGGCUGGGAGCCAAA__GUCCCUGGCCAGAACC .......((((((((((...............................(((((.(((.....))).)))))(((.....))).....))))))))))...... (-30.59 = -30.95 + 0.36)

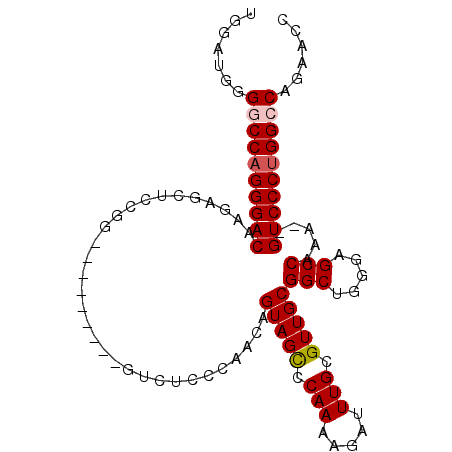
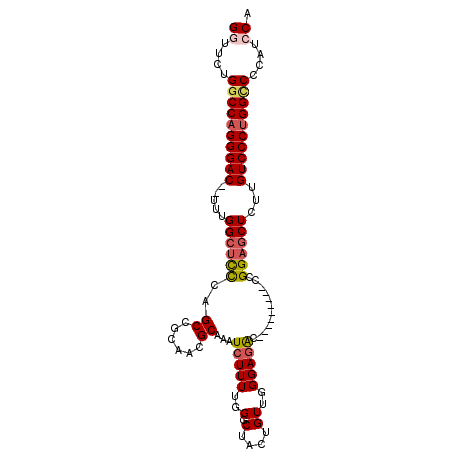
| Location | 12,635,649 – 12,635,741 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 86.78 |
| Mean single sequence MFE | -41.72 |
| Consensus MFE | -32.56 |
| Energy contribution | -33.20 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.15 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12635649 92 - 22407834 GGUUCUGGCCAGGGAC--UUUGGCUCCCAGCCGCAACGCAAAUCUUUUGGGCUACUGUUGGGAGAC---------CCGGAGCUCUUGUCCCUGGCCCCAUCCA ((....((((((((((--...((((((.((((.(((..........)))))))......((....)---------).))))))...))))))))))....)). ( -46.00) >DroSec_CAF1 16666 91 - 1 GGUUCUGCCCAGGGAC--UUUGGCUCCCAGCCGCAACGCAAAUCUUUUGGACUACUGUUGGGAGAC---------CCGGAGCUCUUGUCCCUGGCCCCAU-CA (((...(((.((((((--...((((((..((......))...(((((..(.(....))..))))).---------..))))))...)))))))))...))-). ( -33.40) >DroSim_CAF1 17075 92 - 1 GGUUCUGGCCAGGGAC--UUUGGCUCCCAGCCGCAACGCAAAUCUUUUGGGCUACUGUUGGGAGAC---------CCGGAGCUCUUGUCCCUGGCCCCAUCCA ((....((((((((((--...((((((.((((.(((..........)))))))......((....)---------).))))))...))))))))))....)). ( -46.00) >DroEre_CAF1 16785 92 - 1 GGUUCUGGCCAGGGAC--UUUGGCUCCCAGCCGCAACGCAAAUCUUUUGGGCUACUGUUGGGAGAC---------CCGGACCUCAUGUCCCUGGCCCCAUCCA ((....((((((((((--(((((((((((((.(...(.(((.....))).)...).))))))))..---------)))))......))))))))))....)). ( -40.90) >DroYak_CAF1 17314 100 - 1 GGCUCUGGCCAGGGAC--UUUGGCUCCCAGCCGCAACGCAAAUCUUUUGGGCUACUGUUGGGAGACCCGGAGA-CCCGGAGCUCUUGUCCCUGGCCCCAUCCA ((....((((((((((--...((((((..((......)).........(((((.(((..((....))))))).-)))))))))...))))))))))....)). ( -49.10) >DroAna_CAF1 11479 103 - 1 GUUUCUGGCCUGGGACUCUUUGGCUUUGGGCCGCAACGCAAAUCUUUUGGGCUACUGUUGGGAUGUAUGUGGCUGCUGGUGCUACUGUCCCUGGUCACAGCCA (((..(((((.(((((....((((.(..(((((((..(((..(((..(((....)))..))).))).))))))..)..).))))..))))).))))).))).. ( -34.90) >consensus GGUUCUGGCCAGGGAC__UUUGGCUCCCAGCCGCAACGCAAAUCUUUUGGGCUACUGUUGGGAGAC_________CCGGAGCUCUUGUCCCUGGCCCCAUCCA ((....((((((((((.....((((((..((......))...(((((..(.(....))..)))))............))))))...))))))))))....)). (-32.56 = -33.20 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:45 2006