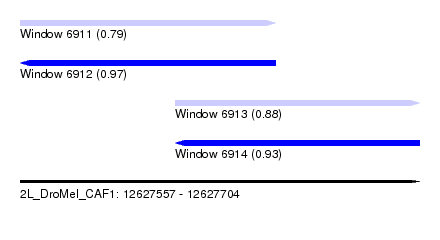
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,627,557 – 12,627,704 |
| Length | 147 |
| Max. P | 0.969716 |

| Location | 12,627,557 – 12,627,651 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 79.85 |
| Mean single sequence MFE | -23.40 |
| Consensus MFE | -16.39 |
| Energy contribution | -15.93 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.790658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
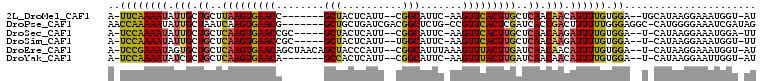
>2L_DroMel_CAF1 12627557 94 + 22407834 A-UUCAAAAUAUUGCUGCUUAAGUGAACC-------GCUACUCAUU--CGGCAUUC-AAGUUCACUUGCUCAACAACAUUUUGUGGA--UGCAUAAGGAAAUGGU-AU .-..((((((.(((.((..(((((((((.-------(((.......--.)))....-..)))))))))..)).))).))))))....--................-.. ( -16.60) >DroPse_CAF1 8449 99 + 1 AACCAAAAAUAUUGCUAAUCAAGUGAACG-------GCUGCUGAUCGACGGCUCUG-CCGUUCACUCGAUCACCGACUUUUUUGGGAGGC-CAUGGGGAAAUCGAUAG ..(((((((..(((...(((.((((((((-------((.((((.....))))...)-))))))))).)))...)))..)))))))((..(-(....))...))..... ( -36.30) >DroSec_CAF1 8504 95 + 1 A-UCCAAAAUAUUGCUGCUCAAGUGAACCGC-----GCUACUCAUU--CGGCAUUC-AAGUUCACUUGCUCAACAAGAUUUUGUGGA--U-CAUAAGGAAAUGGA-UU (-((((.....(((.((..(((((((((...-----(((.......--.)))....-..)))))))))..)).)))((((.....))--)-).........))))-). ( -20.90) >DroSim_CAF1 8589 95 + 1 A-UCCAAAAUAUUGCUGCUCAAGUGAACCGC-----GCUACUCAUU--UGGCAUUC-AAGUUCACUUGCUCAACAAGAUUUUGUGGA--U-CAUAAGGAAAUGGU-UU .-..((((((.(((.((..(((((((((...-----((((......--))))....-..)))))))))..)).))).)))))).(((--(-(((......)))))-)) ( -23.00) >DroEre_CAF1 8513 101 + 1 A-UCCGAAAUAGUGCUGCUCAAGUGAACAGCUAACAGCUACCCAUU--CGGCAUUUAAAGUUUACUUGAUCAACAACAUUUUGUGGA--U-CAUAAGGAAAUGGU-AU .-((((((((..((.((.(((((((((((((.....)))..((...--.))........)))))))))).)).))..)))))).))(--(-(((......)))))-.. ( -23.90) >DroYak_CAF1 8694 93 + 1 A-UCCAAAAUAUCGCUGCUCAAGUGAACA-------GCCACUCAUU--CGGCAUUC-AAGUUUACUUGAUCAACAACAUUUUGUGGA--U-CAUAAGGAAUUGGU-AU .-......(((((((((....((((....-------..))))....--))))....-.(((((...(((((.((((....)))).))--)-))....))))))))-)) ( -19.70) >consensus A_UCCAAAAUAUUGCUGCUCAAGUGAACC_______GCUACUCAUU__CGGCAUUC_AAGUUCACUUGAUCAACAACAUUUUGUGGA__U_CAUAAGGAAAUGGU_AU ..((((((((.(((.((..(((((((((........(((..........))).......)))))))))..)).))).)))))).))...................... (-16.39 = -15.93 + -0.47)
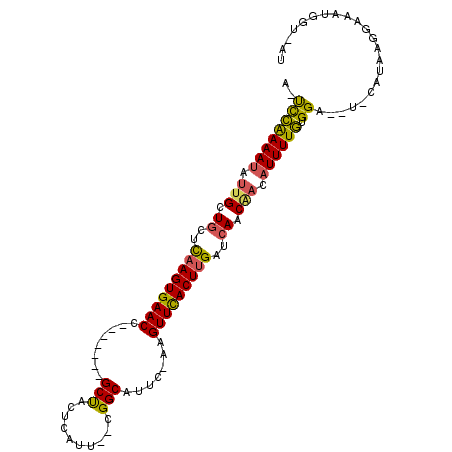

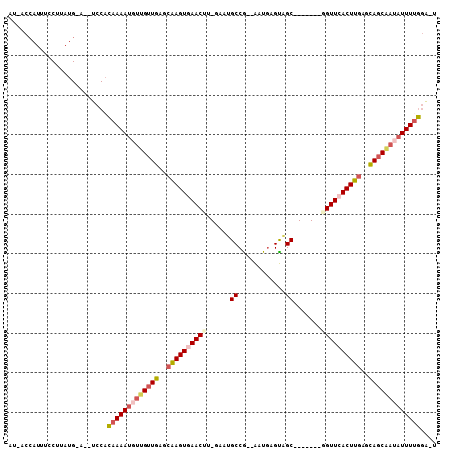
| Location | 12,627,557 – 12,627,651 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 79.85 |
| Mean single sequence MFE | -28.93 |
| Consensus MFE | -21.03 |
| Energy contribution | -22.20 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12627557 94 - 22407834 AU-ACCAUUUCCUUAUGCA--UCCACAAAAUGUUGUUGAGCAAGUGAACUU-GAAUGCCG--AAUGAGUAGC-------GGUUCACUUAAGCAGCAAUAUUUUGAA-U ..-................--....(((((((((((((...(((((((((.-...(((..--.....)))..-------)))))))))...)))))))))))))..-. ( -26.70) >DroPse_CAF1 8449 99 - 1 CUAUCGAUUUCCCCAUG-GCCUCCCAAAAAAGUCGGUGAUCGAGUGAACGG-CAGAGCCGUCGAUCAGCAGC-------CGUUCACUUGAUUAGCAAUAUUUUUGGUU .................-.....(((((((.((.(.(((((((((((((((-(...((.(.....).)).))-------)))))))))))))).).)).))))))).. ( -35.20) >DroSec_CAF1 8504 95 - 1 AA-UCCAUUUCCUUAUG-A--UCCACAAAAUCUUGUUGAGCAAGUGAACUU-GAAUGCCG--AAUGAGUAGC-----GCGGUUCACUUGAGCAGCAAUAUUUUGGA-U ..-..............-(--((((..((((.((((((..((((((((((.-...(((..--.....)))..-----..))))))))))..)))))).))))))))-) ( -28.70) >DroSim_CAF1 8589 95 - 1 AA-ACCAUUUCCUUAUG-A--UCCACAAAAUCUUGUUGAGCAAGUGAACUU-GAAUGCCA--AAUGAGUAGC-----GCGGUUCACUUGAGCAGCAAUAUUUUGGA-U ..-..............-(--((((..((((.((((((..((((((((((.-...((((.--...).)))..-----..))))))))))..)))))).))))))))-) ( -29.30) >DroEre_CAF1 8513 101 - 1 AU-ACCAUUUCCUUAUG-A--UCCACAAAAUGUUGUUGAUCAAGUAAACUUUAAAUGCCG--AAUGGGUAGCUGUUAGCUGUUCACUUGAGCAGCACUAUUUCGGA-U ..-......(((...((-(--((.((((....)))).)))))(((.(((......((((.--....))))...))).(((((((....)))))))))).....)))-. ( -25.80) >DroYak_CAF1 8694 93 - 1 AU-ACCAAUUCCUUAUG-A--UCCACAAAAUGUUGUUGAUCAAGUAAACUU-GAAUGCCG--AAUGAGUGGC-------UGUUCACUUGAGCAGCGAUAUUUUGGA-U ..-..............-(--((((..(((((((((((.((((((.(((..-....((((--......))))-------.))).)))))).)))))))))))))))-) ( -27.90) >consensus AU_ACCAUUUCCUUAUG_A__UCCACAAAAUGUUGUUGAGCAAGUGAACUU_GAAUGCCG__AAUGAGUAGC_______GGUUCACUUGAGCAGCAAUAUUUUGGA_U .........................(((((((((((((..((((((((((......((............)).......))))))))))..))))))))))))).... (-21.03 = -22.20 + 1.17)


| Location | 12,627,614 – 12,627,704 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 81.10 |
| Mean single sequence MFE | -21.80 |
| Consensus MFE | -13.15 |
| Energy contribution | -13.05 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12627614 90 + 22407834 CUCAACAACAUUUUGUGGAUGCAUAAGGAAAUGGUAUUUAUUUGCCU-UG---AUAAGCAAAUGAAUUCGACUUCUUUAUAAGUCUUUUUGGCG ....((((....))))((((..((((((((.(.(.((((((((((..-..---....)))))))))).).).))))))))..))))........ ( -19.90) >DroSec_CAF1 8563 89 + 1 CUCAACAAGAUUUUGUGGAU-CAUAAGGAAAUGGAUUUUAUUUGCCU-UG---AUAAGCAAAUAAAUUCGACUUCUUUAUAAUUAUUUUUGUCG ........(((......(((-.((((((((.(.((.(((((((((..-..---....))))))))).)).).)))))))).)))......))). ( -20.80) >DroSim_CAF1 8648 89 + 1 CUCAACAAGAUUUUGUGGAU-CAUAAGGAAAUGGUUUUUAUUUGCCU-UG---AUAAGCAAAUAAAUUCGACUUCUUUAUAAUUCUUUUUGGCG .....(((((......((((-.((((((((.(.(..(((((((((..-..---....)))))))))..).).)))))))).)))).)))))... ( -20.40) >DroEre_CAF1 8578 88 + 1 AUCAACAACAUUUUGUGGAU-CAUAAGGAAAUGGUAUUUAUUUGCCU-UG---AUGGGCA-AUAAAUUCGGCUUCUUUAUGAGUCUUUGUGGCU .((.((((....)))).))(-(((((((((.(((.(((((((.((((-..---..)))))-)))))))))..))))))))))(((.....))). ( -24.50) >DroYak_CAF1 8751 89 + 1 AUCAACAACAUUUUGUGGAU-CAUAAGGAAUUGGUAUUUAUUUGCCU-UG---AUAAGCAAAUAAAUUCGGCUUUUUUAUGAUGCUUUGUGGCG ........(((.....(.((-(((((((((((((.((((((((((..-..---....)))))))))))))).))))))))))).)...)))... ( -22.40) >DroAna_CAF1 3254 89 + 1 AUAACCAACAUUUUUGAGAU-CACAGGGAAAUGGAAUU-GCUUGCCAAUAAUUAUUUUCAAAUAAAUUCCAUUUCUCUGUCAGUAUA--UGGC- ....(((.((....))....-.((((((((((((((((-..(((..(((....)))..)))...)))))))))))))))).......--))).- ( -22.80) >consensus AUCAACAACAUUUUGUGGAU_CAUAAGGAAAUGGUAUUUAUUUGCCU_UG___AUAAGCAAAUAAAUUCGACUUCUUUAUAAGUCUUUUUGGCG .((.((((....)))).))...((((((((.(.(.((((((((((............)))))))))).).).)))))))).............. (-13.15 = -13.05 + -0.10)


| Location | 12,627,614 – 12,627,704 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 81.10 |
| Mean single sequence MFE | -18.31 |
| Consensus MFE | -12.73 |
| Energy contribution | -13.82 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12627614 90 - 22407834 CGCCAAAAAGACUUAUAAAGAAGUCGAAUUCAUUUGCUUAU---CA-AGGCAAAUAAAUACCAUUUCCUUAUGCAUCCACAAAAUGUUGUUGAG .((......(((((......)))))..(((.((((((((..---..-)))))))).))).............)).((.((((....)))).)). ( -14.90) >DroSec_CAF1 8563 89 - 1 CGACAAAAAUAAUUAUAAAGAAGUCGAAUUUAUUUGCUUAU---CA-AGGCAAAUAAAAUCCAUUUCCUUAUG-AUCCACAAAAUCUUGUUGAG ((((((.....(((((((.(((((.((.(((((((((((..---..-))))))))))).)).))))).)))))-))..........)))))).. ( -21.76) >DroSim_CAF1 8648 89 - 1 CGCCAAAAAGAAUUAUAAAGAAGUCGAAUUUAUUUGCUUAU---CA-AGGCAAAUAAAAACCAUUUCCUUAUG-AUCCACAAAAUCUUGUUGAG ...(((.(((((((((((.(((((.(..(((((((((((..---..-)))))))))))..).))))).)))))-))........)))).))).. ( -19.20) >DroEre_CAF1 8578 88 - 1 AGCCACAAAGACUCAUAAAGAAGCCGAAUUUAU-UGCCCAU---CA-AGGCAAAUAAAUACCAUUUCCUUAUG-AUCCACAAAAUGUUGUUGAU ............((((((.((((..(.((((((-((((...---..-.))).))))))).)..)))).)))))-)((.((((....)))).)). ( -16.10) >DroYak_CAF1 8751 89 - 1 CGCCACAAAGCAUCAUAAAAAAGCCGAAUUUAUUUGCUUAU---CA-AGGCAAAUAAAUACCAAUUCCUUAUG-AUCCACAAAAUGUUGUUGAU .((......)).((((((..((...(.((((((((((((..---..-)))))))))))).)...))..)))))-)((.((((....)))).)). ( -17.00) >DroAna_CAF1 3254 89 - 1 -GCCA--UAUACUGACAGAGAAAUGGAAUUUAUUUGAAAAUAAUUAUUGGCAAGC-AAUUCCAUUUCCCUGUG-AUCUCAAAAAUGUUGGUUAU -((((--.......((((.(((((((((((..((((..(((....)))..)))).-))))))))))).))))(-....)........))))... ( -20.90) >consensus CGCCAAAAAGAAUUAUAAAGAAGUCGAAUUUAUUUGCUUAU___CA_AGGCAAAUAAAUACCAUUUCCUUAUG_AUCCACAAAAUGUUGUUGAG .............(((((.(((((.(.((((((((((((........)))))))))))).).))))).)))))..((.((((....)))).)). (-12.73 = -13.82 + 1.09)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:39 2006