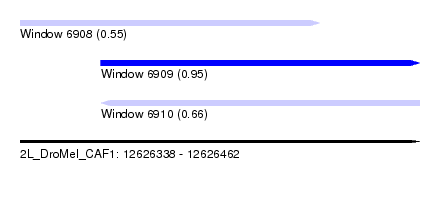
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,626,338 – 12,626,462 |
| Length | 124 |
| Max. P | 0.949853 |

| Location | 12,626,338 – 12,626,431 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 74.33 |
| Mean single sequence MFE | -28.58 |
| Consensus MFE | -16.25 |
| Energy contribution | -16.67 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12626338 93 + 22407834 UCACCCCAGCAGUUGCACCAUCA--CCACCACCAUCCACUGCAAAUCACGCCGCCCAGCUCGGCGGCCAGUUUGAAAUUGAGU---GGUAUGCUGACG ......((((((.((......))--.)(((((((.......(((((...((((((......))))))..)))))....)).))---))).)))))... ( -27.60) >DroVir_CAF1 64445 90 + 1 UCGCCGCAGUCGCAGCAG-----GCGUCGCAUCAUCCGCUGCAGAUAACGCCGCCCAAUUCGGCGGCCAGCCUUAAGCUGAGC---GGCAUGCUUACG ..(((((.((((((((.(-----..(......)..).))))).)))...((((((......))))))((((.....)))).))---)))......... ( -38.40) >DroPse_CAF1 7372 84 + 1 UCCCCGCAGCAGCUCCA--------------CCAUCCGCUCCAGAUCACGCCGCCCAGCUCGGCGGCCAGCAUCAAGAUGAGCGGCGGCAUGCUGACC ......((((((((...--------------....((((((..(((...((((((......))))))....))).....)))))).))).)))))... ( -30.10) >DroSec_CAF1 7289 93 + 1 UCACCCCAGCAGAUGCACCACCA--CCACCCCCAUCCACUGCAAAUCACGCCGCCCAGCUCAGCGGCCAGUUUGAAAUUGAGC---GGUAUGCUGACG ......(((((((((........--.......)))).(((((.......(((((........)))))((((.....)))).))---))).)))))... ( -23.56) >DroYak_CAF1 7437 93 + 1 UCACCCCACCAGAUGCACCACCA--CCACCAUCACCCACUGCAGAUCACUCCGCCCAGCUCGGCGGCCAGUCUGAAACUGAGU---GGAAUGCUGACG .....((((((((((........--....))))........(((((....(((((......)))))...)))))....)).))---)).......... ( -21.70) >DroPer_CAF1 7614 84 + 1 UCCCCGCAGCAGCUCCA--------------CCAUCCGCUCCAGAUCACGCCGCCCAGCUCGGCGGCCAGCAUCAAGAUGAGCGGCGGCAUGCUGACC ......((((((((...--------------....((((((..(((...((((((......))))))....))).....)))))).))).)))))... ( -30.10) >consensus UCACCCCAGCAGCUGCAC_______CCACCACCAUCCACUGCAGAUCACGCCGCCCAGCUCGGCGGCCAGCAUGAAAAUGAGC___GGCAUGCUGACG ......(((((......................................((((((......))))))((((.....))))..........)))))... (-16.25 = -16.67 + 0.42)


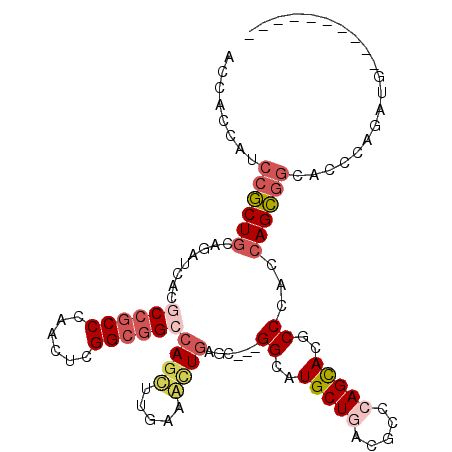
| Location | 12,626,363 – 12,626,462 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 76.13 |
| Mean single sequence MFE | -33.48 |
| Consensus MFE | -23.66 |
| Energy contribution | -24.63 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.949853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12626363 99 + 22407834 ACCACCAUCCACUGCAAAUCACGCCGCCCAGCUCGGCGGCCAGUUUGAAAUUGAGU---GGUAUGCUGACGCCCAGUACGCCCACCAGUGGCACCCAGAUGA--------- ........(((((((((((...((((((......))))))..))))).......((---((..(((((.....)))))..).)))))))))...........--------- ( -33.00) >DroVir_CAF1 64467 95 + 1 CGCAUCAUCCGCUGCAGAUAACGCCGCCCAAUUCGGCGGCCAGCCUUAAGCUGAGC---GGCAUGCUUACGCCCAGCACGCCCACCAGCGGCACCCAC------------- ........(((((((.......((((((......))))))((((.....)))).))---(((.((((.......)))).)))....))))).......------------- ( -35.30) >DroPse_CAF1 7389 97 + 1 ----CCAUCCGCUCCAGAUCACGCCGCCCAGCUCGGCGGCCAGCAUCAAGAUGAGCGGCGGCAUGCUGACCCCCAGCACCCCCACCAGCGGCACGCAGAUG---------- ----.(((((((((..(((...((((((......))))))....))).....)))))(((((.(((((.................))))))).))).))))---------- ( -34.03) >DroYak_CAF1 7462 99 + 1 ACCAUCACCCACUGCAGAUCACUCCGCCCAGCUCGGCGGCCAGUCUGAAACUGAGU---GGAAUGCUGACGCCCAGCACGCCCACCAGUGGCACCCAGAUGA--------- ..((((..(((((((((((....(((((......)))))...))))).......((---((..(((((.....)))))...))))))))))......)))).--------- ( -35.60) >DroMoj_CAF1 60853 100 + 1 --CAUCAGCCGCUGCAGAUAACGCCGCCUAAUUCGGCAGCCAGCUUGAAGCUGAGC---GGCAUGCUAACGCCCAGCACGCCCACCAGCCACUCGCACACCCAUG------ --.....((.((((........((((((......))).((((((.....)))).))---)))..((....)).))))..((......)).....)).........------ ( -28.90) >DroAna_CAF1 2079 108 + 1 ACCACCUGCCGCUGCAGAUCACACCCCCCAACUCGGCGGCCAGCUUGAAACUAAGC---GGAAUGCUGACUCCCAGCACACCCACCAGCGGCACCCAGAUGCAGAGCGGCU .......(((((((((((..............))((..(((.(((((....)))))---((..(((((.....)))))..)).......)))..))...)))..)))))). ( -34.04) >consensus ACCACCAUCCGCUGCAGAUCACGCCGCCCAACUCGGCGGCCAGCUUGAAACUGAGC___GGCAUGCUGACGCCCAGCACGCCCACCAGCGGCACCCAGAUG__________ ........((((((........((((((......))))))((((.....))))......((..(((((.....)))))..))...)))))).................... (-23.66 = -24.63 + 0.98)


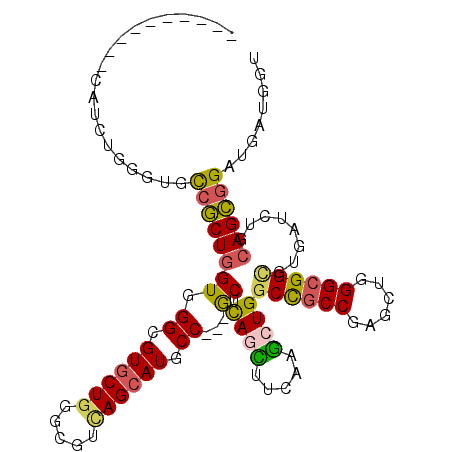
| Location | 12,626,363 – 12,626,462 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 76.13 |
| Mean single sequence MFE | -43.30 |
| Consensus MFE | -30.56 |
| Energy contribution | -30.12 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12626363 99 - 22407834 ---------UCAUCUGGGUGCCACUGGUGGGCGUACUGGGCGUCAGCAUACC---ACUCAAUUUCAAACUGGCCGCCGAGCUGGGCGGCGUGAUUUGCAGUGGAUGGUGGU ---------..........((((((.((.(((((.....))))).))...((---((((((..(((.....((((((......)))))).))).))).)))))..)))))) ( -37.80) >DroVir_CAF1 64467 95 - 1 -------------GUGGGUGCCGCUGGUGGGCGUGCUGGGCGUAAGCAUGCC---GCUCAGCUUAAGGCUGGCCGCCGAAUUGGGCGGCGUUAUCUGCAGCGGAUGAUGCG -------------(((....((((((...((((((((.......))))))))---...((((.....))))((((((......))))))........))))))....))). ( -46.10) >DroPse_CAF1 7389 97 - 1 ----------CAUCUGCGUGCCGCUGGUGGGGGUGCUGGGGGUCAGCAUGCCGCCGCUCAUCUUGAUGCUGGCCGCCGAGCUGGGCGGCGUGAUCUGGAGCGGAUGG---- ----------(((((((...((.......((.((((((.....)))))).))((((((((.((((..((.....)))))).)))))))).......)).))))))).---- ( -46.70) >DroYak_CAF1 7462 99 - 1 ---------UCAUCUGGGUGCCACUGGUGGGCGUGCUGGGCGUCAGCAUUCC---ACUCAGUUUCAGACUGGCCGCCGAGCUGGGCGGAGUGAUCUGCAGUGGGUGAUGGU ---------(((((......(((((((((((.((((((.....)))))))))---)).......(((((...(((((......)))))...).))))))))))..))))). ( -39.90) >DroMoj_CAF1 60853 100 - 1 ------CAUGGGUGUGCGAGUGGCUGGUGGGCGUGCUGGGCGUUAGCAUGCC---GCUCAGCUUCAAGCUGGCUGCCGAAUUAGGCGGCGUUAUCUGCAGCGGCUGAUG-- ------(((.((((((((..((((.(((((.(((((((.....)))))))))---)))((((.....))))((((((......))))))))))..)))).).))).)))-- ( -43.10) >DroAna_CAF1 2079 108 - 1 AGCCGCUCUGCAUCUGGGUGCCGCUGGUGGGUGUGCUGGGAGUCAGCAUUCC---GCUUAGUUUCAAGCUGGCCGCCGAGUUGGGGGGUGUGAUCUGCAGCGGCAGGUGGU .((((((((.((.((.(((((((((((((((.((((((.....)))))))))---)))........))).)).)))).)).)).)))))...((((((....))))))))) ( -46.20) >consensus __________CAUCUGGGUGCCGCUGGUGGGCGUGCUGGGCGUCAGCAUGCC___GCUCAGCUUCAAGCUGGCCGCCGAGCUGGGCGGCGUGAUCUGCAGCGGAUGAUGGU ....................((((((((.((.((((((.....)))))).))...)).((((.....))))((((((......))))))........))))))........ (-30.56 = -30.12 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:36 2006