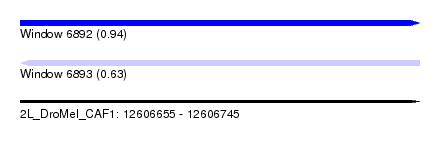
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,606,655 – 12,606,745 |
| Length | 90 |
| Max. P | 0.937761 |

| Location | 12,606,655 – 12,606,745 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.24 |
| Mean single sequence MFE | -24.88 |
| Consensus MFE | -16.36 |
| Energy contribution | -16.78 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12606655 90 + 22407834 ---------------------------AAGUAAAUUGCA---AUAAUUGUACUAAUUGUUAUUGCAAUUAUUGCUGGCAAUUACCAUGACUAAUGUGGGGCAAAAUGUUUGUGCAUAUUG ---------------------------.(((((((((((---(((((.((....)).)))))))))))..))))).((.....((((.......)))).((((.....))))))...... ( -19.40) >DroPse_CAF1 232506 101 + 1 AC------------ACACAUAC-AGCCAGGUAAAUUGCA---AUAAUUGUACUAAUUGUUAUUGCAAUUAUUGUUGGCAAUUACCAUGACUAAUGUGGGGCAAAAUGUUUGUGAAUG--- ..------------.((((.((-((((((...(((((((---(((((.((....)).))))))))))))....))))).....((((.......)))).......))).))))....--- ( -28.10) >DroWil_CAF1 266347 102 + 1 CA------------AAACA------UCACGUCAAUUGCAAAAAUAAUUGUACUAAUUGUUUAUGCAAUUAUUGUUGGUAAUUACCAUGACUAAUGUGGGGCAAAAUGUUUAUGAAUAAUA ..------------(((((------(...(((((((((...((((((((((.(((....)))))))))))))....)))))).((((.......)))))))...)))))).......... ( -22.00) >DroYak_CAF1 177703 113 + 1 AC----AUACUUACAUAUAUACGAGCCAAGUAAAUUGCA---AUAAUUGUACUAAUUGUUAUUGCAAUUAUUGCUGGCAAUUACCAUGACUAAUGUGGGGCAAAAUGUUUGUGCAUAUUG ..----.(((..((((........((((.((((((((((---(((((.((....)).)))))))))))..)))))))).....((((.......))))......))))..)))....... ( -27.30) >DroAna_CAF1 172462 115 + 1 ACGAAAAAAUCC--ACAAACAUAAGCUACGUAAAUUGCA---AUAAUUGUACUAAUUGUUAUUGCAAUUAUUGCUGGCAAUUACCAUGACUAAUGUGGGGCAAAAUGUUUAUGAAUAAUA ...........(--(.((((((..((((.((((((((((---(((((.((....)).)))))))))))..)))))))).....((((.......))))......)))))).))....... ( -25.40) >DroPer_CAF1 226017 101 + 1 AC------------ACACAUAC-UGCUAGGUAAAUUGCA---AUAAUUGUACUAAUUGUUAUUGCAAUUAUUGCUGGCAAUUACCAUGACUAAUGUGGGGCAAAAUGUUUGUGAAUG--- ..------------.((((.((-((((((...(((((((---(((((.((....)).))))))))))))....))))))....((((.......))))........)).))))....--- ( -27.10) >consensus AC____________ACACAUAC_AGCCAAGUAAAUUGCA___AUAAUUGUACUAAUUGUUAUUGCAAUUAUUGCUGGCAAUUACCAUGACUAAUGUGGGGCAAAAUGUUUGUGAAUA_U_ ........................(((.....((((((....(((((((((.((.....)).))))))))).....)))))).((((.......)))))))................... (-16.36 = -16.78 + 0.42)


| Location | 12,606,655 – 12,606,745 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.24 |
| Mean single sequence MFE | -21.62 |
| Consensus MFE | -9.11 |
| Energy contribution | -8.75 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.632681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12606655 90 - 22407834 CAAUAUGCACAAACAUUUUGCCCCACAUUAGUCAUGGUAAUUGCCAGCAAUAAUUGCAAUAACAAUUAGUACAAUUAU---UGCAAUUUACUU--------------------------- (((.(((......))).)))..............((((....))))((((((((((...((.....))...)))))))---))).........--------------------------- ( -14.30) >DroPse_CAF1 232506 101 - 1 ---CAUUCACAAACAUUUUGCCCCACAUUAGUCAUGGUAAUUGCCAACAAUAAUUGCAAUAACAAUUAGUACAAUUAU---UGCAAUUUACCUGGCU-GUAUGUGU------------GU ---....................(((((((((((.((((((((....))))(((((((((((............))))---))))))))))))))))-).))))).------------.. ( -24.10) >DroWil_CAF1 266347 102 - 1 UAUUAUUCAUAAACAUUUUGCCCCACAUUAGUCAUGGUAAUUACCAACAAUAAUUGCAUAAACAAUUAGUACAAUUAUUUUUGCAAUUGACGUGA------UGUUU------------UG ........................(((((((((.((((....)))).....(((((((.(((.((((.....)))).))).)))))))))).)))------)))..------------.. ( -17.80) >DroYak_CAF1 177703 113 - 1 CAAUAUGCACAAACAUUUUGCCCCACAUUAGUCAUGGUAAUUGCCAGCAAUAAUUGCAAUAACAAUUAGUACAAUUAU---UGCAAUUUACUUGGCUCGUAUAUAUGUAAGUAU----GU ..((((((.........(((((..((....))...)))))..(((((....(((((((((((............))))---)))))))...)))))..))))))..........----.. ( -23.30) >DroAna_CAF1 172462 115 - 1 UAUUAUUCAUAAACAUUUUGCCCCACAUUAGUCAUGGUAAUUGCCAGCAAUAAUUGCAAUAACAAUUAGUACAAUUAU---UGCAAUUUACGUAGCUUAUGUUUGU--GGAUUUUUUCGU ....((((((((((((...((..(..........((((....))))((((((((((...((.....))...)))))))---))).......)..))..))))))))--))))........ ( -28.10) >DroPer_CAF1 226017 101 - 1 ---CAUUCACAAACAUUUUGCCCCACAUUAGUCAUGGUAAUUGCCAGCAAUAAUUGCAAUAACAAUUAGUACAAUUAU---UGCAAUUUACCUAGCA-GUAUGUGU------------GU ---.....(((.((((.((((.............((((....))))((((((((((...((.....))...)))))))---)))..........)))-).)))).)------------)) ( -22.10) >consensus _A_UAUUCACAAACAUUUUGCCCCACAUUAGUCAUGGUAAUUGCCAGCAAUAAUUGCAAUAACAAUUAGUACAAUUAU___UGCAAUUUACCUGGCU_GUAUGUGU____________GU .................((((.........((..((((....))))))..((((((......))))))..............)))).................................. ( -9.11 = -8.75 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:20 2006