
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,574,311 – 12,574,422 |
| Length | 111 |
| Max. P | 0.867650 |

| Location | 12,574,311 – 12,574,422 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.87 |
| Mean single sequence MFE | -23.57 |
| Consensus MFE | -17.77 |
| Energy contribution | -17.77 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12574311 111 - 22407834 CUCGAA---AAGGCAAAAACUGUUUAUGCAUUUUGUGCA-GCGAGUGCAACAAAUUUAAUUGUACC-GAUAUAGACAUGCAUUGGCAGUAAAAACCAA-AACUAAUAAU---ACUGGCGA ......---..((.....(((((..((((((((((((..-.((.((((((.........)))))))-).)))))).))))))..))))).....))..-..........---........ ( -24.20) >DroPse_CAF1 186868 112 - 1 GUUGCAACAACAGCGGCAACUGUUUAUGCAUUUUGUGCA-GCGAGUGCAACAAAUUAAAUUGUGCC-GAU--AGACAUGCAUUGGCAGUAAAAGCCAAAAACUAAUAAU---ACUCC-AC ((((......))))(((.(((((..(((((((((((...-.((.(..(((.........)))..))-)))--))).))))))..)))))....))).............---.....-.. ( -25.90) >DroGri_CAF1 198841 108 - 1 ACUGCAACUGCAACUGAAACUGUUUAUGCAUUUUGUGCAGGCGAGUGCAACAAAUUAAAUUGUACU-UUU--AGACAUGCAUUGGCAGUAAAACACAC-ACAAAACAGA---AAU----- (((((...((((.((((....((...(((((...))))).))((((((((.........)))))))-)))--))...))))...))))).........-..........---...----- ( -22.60) >DroWil_CAF1 196956 113 - 1 --UGCAA--CUUGGGCAAACUGUUUAUGCAUUUUGUGCA-GCGAGUGCAACAAAUUUAAUUGUAGUAGAU-CAGACAUGCAUUGGCAGUAAAAAUUAU-AACUAAAAACUUAACAAACAA --(((..--..((.(((((.(((....))).))))).))-((.((((((((((......)))).((....-...)).)))))).)).)))........-..................... ( -20.50) >DroAna_CAF1 140505 105 - 1 C----A---GCCGCAGCAACUGUUUAUGCAUUUUGUGCA-GCGAGUGCAACAACUUUAAUUGUGCC-GAU--AGACAUGCAUUGGCAGUAAAAACCAA-AACUAAUAAC---ACUCAAGC .----.---((.(((.(((.(((....)))..)))))).-))(((((..((((......))))(((-(((--........))))))............-.........)---)))).... ( -22.30) >DroPer_CAF1 183944 112 - 1 GUUGCAACAACAGCGGCAACUGUUUAUGCAUUUUGUGCA-GCGAGUGCAACAAAUUUAAUUGUGCC-GAU--AGACAUGCAUUGGCAGUAAAAGCCAAAAACUAAUAAU---ACUCC-AC ((((......))))(((.(((((..(((((((((((...-.((.(..(((.........)))..))-)))--))).))))))..)))))....))).............---.....-.. ( -25.90) >consensus CUUGCAA__ACAGCGGAAACUGUUUAUGCAUUUUGUGCA_GCGAGUGCAACAAAUUUAAUUGUACC_GAU__AGACAUGCAUUGGCAGUAAAAACCAA_AACUAAUAAU___ACUCA_AC ..................(((((..((((((..((..(......)..))((((......)))).............))))))..)))))............................... (-17.77 = -17.77 + 0.00)

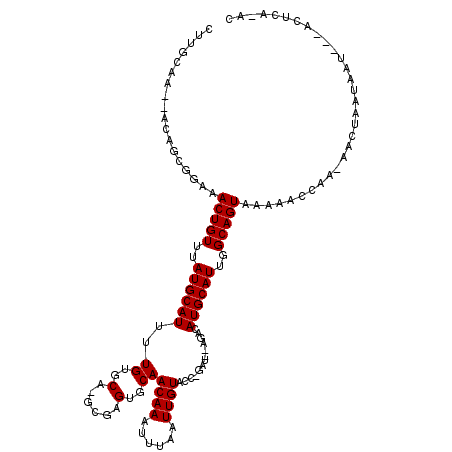
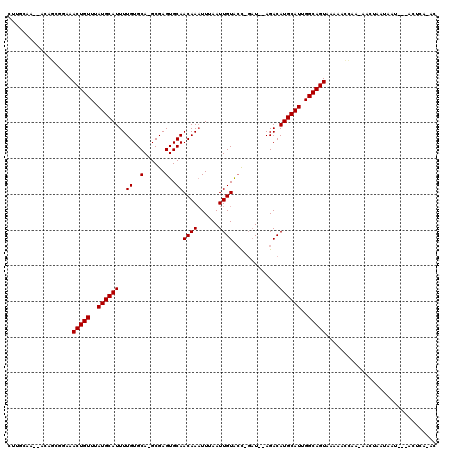
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:08 2006