| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,567,742 – 12,567,839 |
| Length | 97 |
| Max. P | 0.793100 |

| Location | 12,567,742 – 12,567,839 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 80.95 |
| Mean single sequence MFE | -20.37 |
| Consensus MFE | -13.84 |
| Energy contribution | -14.27 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12567742 97 + 22407834 UGAUUCGCAGGAAAAUUUCCUUGGCCAUGCCGUUUGACCAUUUCCACUCUCUCCCCAUCGAGCUGCUUUUCCGAGCUGCUUUUCCGGCCUUUGCCAU .........((((((.(..(((((....((.((((((....................)))))).))....)))))..).))))))(((....))).. ( -22.05) >DroSec_CAF1 132914 92 + 1 UGAUUCGCAGGAAAAUUUCCUUGGCCGUGCCGUUUGACCAUUUCCACUCUCUCCCCU-----CGGCUUUUCCGAGCUGCUUUUCCGGCCUUUGCCAU .........((((((.(..(((((....((((.........................-----))))....)))))..).))))))(((....))).. ( -21.11) >DroSim_CAF1 135589 92 + 1 UGAUUCGCAGGAAAAUUUCCUUGGCCAUGCCGUUUGACCAAUUCCACUCUCUCCCCU-----CGGCCUUUUCGAGCUGCUUUUCCGGCCUUUGCCAU .........((((((.(..(((((....((((.........................-----))))....)))))..).))))))(((....))).. ( -17.91) >DroEre_CAF1 131714 78 + 1 UGAUUCGCAGGAAAAUUUCUUCGG------------UCCAAUUCCACUCCCC--CCG-----CGGCCCAUCUGCGCCGCUUUUCCGGCCUUUGCCAU .........((((((.......((------------.......)).......--..(-----((((.(....).)))))))))))(((....))).. ( -20.40) >consensus UGAUUCGCAGGAAAAUUUCCUUGGCCAUGCCGUUUGACCAAUUCCACUCUCUCCCCU_____CGGCCUUUCCGAGCUGCUUUUCCGGCCUUUGCCAU .........((((((.......((.((.......)).)).......................((((((....)))))).))))))(((....))).. (-13.84 = -14.27 + 0.44)



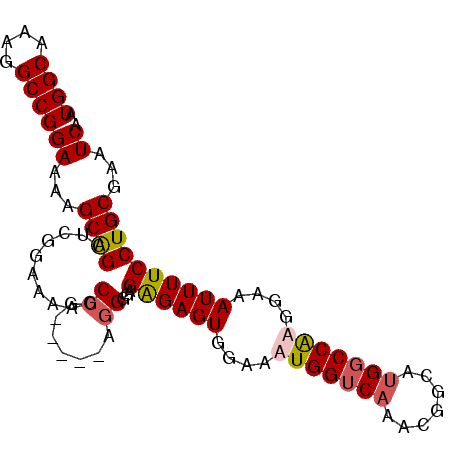
| Location | 12,567,742 – 12,567,839 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 80.95 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -18.26 |
| Energy contribution | -19.07 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
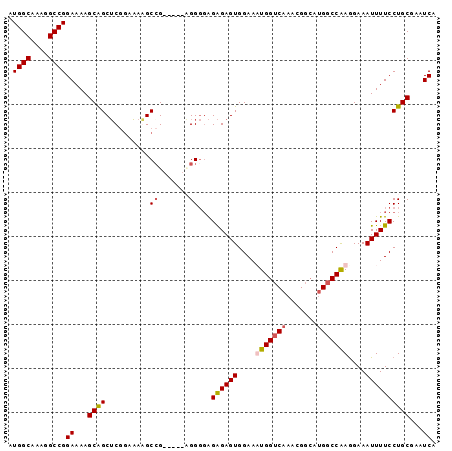
>2L_DroMel_CAF1 12567742 97 - 22407834 AUGGCAAAGGCCGGAAAAGCAGCUCGGAAAAGCAGCUCGAUGGGGAGAGAGUGGAAAUGGUCAAACGGCAUGGCCAAGGAAAUUUUCCUGCGAAUCA .((((....((((....(((.(((......))).))).........((..((....))..))...))))...))))((((.....))))........ ( -24.80) >DroSec_CAF1 132914 92 - 1 AUGGCAAAGGCCGGAAAAGCAGCUCGGAAAAGCCG-----AGGGGAGAGAGUGGAAAUGGUCAAACGGCACGGCCAAGGAAAUUUUCCUGCGAAUCA .((((....((((.........(((((.....)))-----))....((..((....))..))...))))...))))((((.....))))........ ( -27.60) >DroSim_CAF1 135589 92 - 1 AUGGCAAAGGCCGGAAAAGCAGCUCGAAAAGGCCG-----AGGGGAGAGAGUGGAAUUGGUCAAACGGCAUGGCCAAGGAAAUUUUCCUGCGAAUCA .((((....))))((...((((((((.......))-----))....((((((....(((((((.......)))))))....))))))))))...)). ( -27.30) >DroEre_CAF1 131714 78 - 1 AUGGCAAAGGCCGGAAAAGCGGCGCAGAUGGGCCG-----CGG--GGGGAGUGGAAUUGGA------------CCGAAGAAAUUUUCCUGCGAAUCA .((((....))))((.....(((.(....).)))(-----(((--(..((.(((.......------------)))......))..)))))...)). ( -24.20) >consensus AUGGCAAAGGCCGGAAAAGCAGCUCGGAAAAGCCG_____AGGGGAGAGAGUGGAAAUGGUCAAACGGCAUGGCCAAGGAAAUUUUCCUGCGAAUCA .((((....))))((...((((..........((.......))...((((((....(((((((.......)))))))....))))))))))...)). (-18.26 = -19.07 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:07 2006