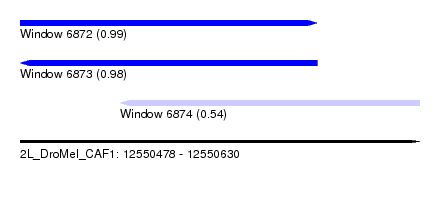
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,550,478 – 12,550,630 |
| Length | 152 |
| Max. P | 0.987195 |

| Location | 12,550,478 – 12,550,591 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 77.59 |
| Mean single sequence MFE | -38.02 |
| Consensus MFE | -26.02 |
| Energy contribution | -26.33 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
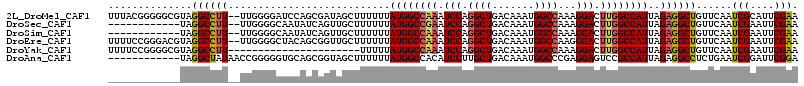
>2L_DroMel_CAF1 12550478 113 + 22407834 UUUACGGGGGCGUAGGCCUU--UUGGGGAUCCAGCGAUAGCUUUUUUAUGGCCAAAUCCAGGCUGACAAAUGGCCAAAGGACUUGGCCAUUAGAGGCUGUUCAAUCGCAUUCGAA ....((((.((((.(((((.--...)))..)).))((((((((((..((((((((.(((.((((.......))))...))).)))))))).)))))))))).....)).)))).. ( -42.10) >DroSec_CAF1 122564 101 + 1 ------------UAGGCCUU--UUGGGGCAAUAUCAGUUGCUUUUUUAUGGCCGAAUCCAGGCUGACAAAUGGCCAAAGGACUUGGCCAUUAGAGGCUGUUCAAUCGAAUUCGAA ------------..((((((--(.((((((((....))))))))...((((((((.(((.((((.......))))...))).)))))))).)))))))......(((....))). ( -36.60) >DroSim_CAF1 125402 101 + 1 ------------UAGGCCUU--UUGGGGCAAUAUCAGUUGCUUUUUUAUGGCCAAAUCCAGGCUGACAAAUGGCCAAAGGACUUGGCCAUUAGAGGCUGUUCAAUCGAAUUCGAA ------------..((((((--(.((((((((....))))))))...((((((((.(((.((((.......))))...))).)))))))).)))))))......(((....))). ( -36.90) >DroEre_CAF1 121605 113 + 1 UUUUCCGGGACGUAGGCCUU--UUGGGGCUACAGCGGUUGCUUUUUUAUGGCCAAAUCCAGGCUGACAAAUGGCCAAGGGACUUGGCCAUUAGAGGCUGUUCAAUCGAAUUCGAA .......((((((((.((..--...)).)))).......((((((..((((((((.(((.((((.......))))...))).)))))))).)))))).))))..(((....))). ( -43.60) >DroYak_CAF1 126166 93 + 1 UUUUCCGGGGCGUAGGCCUU----------------------UUUUUAUGGCCAAAUCCAGGCUGACAAAUGGCCAAAGGACUUGGCCAUUAGAGGCUGUUCAAUCGAAUUCGAA .......((((...((((((----------------------(....((((((((.(((.((((.......))))...))).)))))))).)))))))))))..(((....))). ( -35.00) >DroAna_CAF1 122936 103 + 1 ------------UAGGCUAUAACCGGGGGUGCAGCGGUAGCUUUUUUAUGGCCACAUCCUUGCUGACAAAUGGCCCGAGGAGUCCGCCAUUAGAGGCCUCUGAAUCGGAUUCGGA ------------..((((((...((((((((..((.((((.....)))).))..)))))))).......))))))....(((((((...(((((....)))))..)))))))... ( -33.90) >consensus ____________UAGGCCUU__UUGGGGCUACAGCGGUUGCUUUUUUAUGGCCAAAUCCAGGCUGACAAAUGGCCAAAGGACUUGGCCAUUAGAGGCUGUUCAAUCGAAUUCGAA ..............((((((...........................((((((((.(((.((((.......))))...))).))))))))..))))))......(((....))). (-26.02 = -26.33 + 0.31)
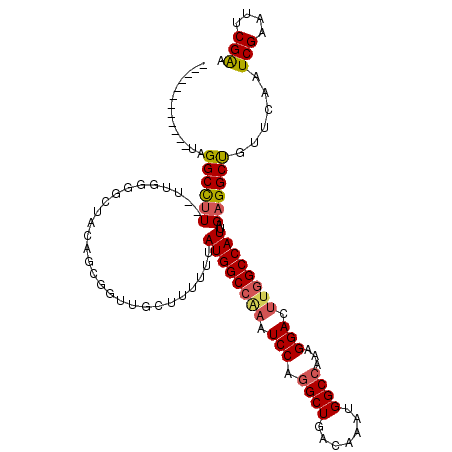
| Location | 12,550,478 – 12,550,591 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 77.59 |
| Mean single sequence MFE | -31.73 |
| Consensus MFE | -21.50 |
| Energy contribution | -22.77 |
| Covariance contribution | 1.26 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.978996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12550478 113 - 22407834 UUCGAAUGCGAUUGAACAGCCUCUAAUGGCCAAGUCCUUUGGCCAUUUGUCAGCCUGGAUUUGGCCAUAAAAAAGCUAUCGCUGGAUCCCCAA--AAGGCCUACGCCCCCGUAAA .......(((((.....(((.....((((((((((((...(((.........))).))))))))))))......))))))))(((....))).--..(((....)))........ ( -35.80) >DroSec_CAF1 122564 101 - 1 UUCGAAUUCGAUUGAACAGCCUCUAAUGGCCAAGUCCUUUGGCCAUUUGUCAGCCUGGAUUCGGCCAUAAAAAAGCAACUGAUAUUGCCCCAA--AAGGCCUA------------ .(((....))).......((((...((((((.(((((...(((.........))).))))).))))))......((((......)))).....--.))))...------------ ( -29.10) >DroSim_CAF1 125402 101 - 1 UUCGAAUUCGAUUGAACAGCCUCUAAUGGCCAAGUCCUUUGGCCAUUUGUCAGCCUGGAUUUGGCCAUAAAAAAGCAACUGAUAUUGCCCCAA--AAGGCCUA------------ .(((....))).......((((...((((((((((((...(((.........))).))))))))))))......((((......)))).....--.))))...------------ ( -33.20) >DroEre_CAF1 121605 113 - 1 UUCGAAUUCGAUUGAACAGCCUCUAAUGGCCAAGUCCCUUGGCCAUUUGUCAGCCUGGAUUUGGCCAUAAAAAAGCAACCGCUGUAGCCCCAA--AAGGCCUACGUCCCGGAAAA .(((....))).......((.....((((((((((((...(((.........))).))))))))))))......))..(((.(((((((....--..)).)))))...))).... ( -35.70) >DroYak_CAF1 126166 93 - 1 UUCGAAUUCGAUUGAACAGCCUCUAAUGGCCAAGUCCUUUGGCCAUUUGUCAGCCUGGAUUUGGCCAUAAAAA----------------------AAGGCCUACGCCCCGGAAAA .(((....)))........((....((((((((((((...(((.........))).)))))))))))).....----------------------..(((....)))..)).... ( -30.50) >DroAna_CAF1 122936 103 - 1 UCCGAAUCCGAUUCAGAGGCCUCUAAUGGCGGACUCCUCGGGCCAUUUGUCAGCAAGGAUGUGGCCAUAAAAAAGCUACCGCUGCACCCCCGGUUAUAGCCUA------------ (((((((...))))....(((......)))))).......((((((..(((......)))))))))........((((..((((......))))..))))...------------ ( -26.10) >consensus UUCGAAUUCGAUUGAACAGCCUCUAAUGGCCAAGUCCUUUGGCCAUUUGUCAGCCUGGAUUUGGCCAUAAAAAAGCAACCGCUGUAGCCCCAA__AAGGCCUA____________ .(((....))).......((((...((((((((((((...(((.........))).)))))))))))).......(....)...............))))............... (-21.50 = -22.77 + 1.26)



| Location | 12,550,516 – 12,550,630 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.07 |
| Mean single sequence MFE | -30.05 |
| Consensus MFE | -23.47 |
| Energy contribution | -24.92 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12550516 114 - 22407834 AG-UCAUAAAUUAUGGGCAAUUAAUAUGUAACACGUUAGA--UUCGAAUGCGAUU-GAACAGCCUCUAAUGGCCAAGUCCUU--UGGCCAUUUGUCAGCCUGGAUUUGGCCAUAAAAAAG ..-......((((.((((..(((((.((((...((.....--..))..)))))))-))...)))).))))((((((((((..--.(((.........))).))))))))))......... ( -32.00) >DroSec_CAF1 122590 114 - 1 AG-UCAUAAAUUAUGGGCAAUUAAUAUGUAACACGUUAGA--UUCGAAUUCGAUU-GAACAGCCUCUAAUGGCCAAGUCCUU--UGGCCAUUUGUCAGCCUGGAUUCGGCCAUAAAAAAG ..-......((((.((((.........(.....)(((...--.(((....)))..-.))).)))).))))((((.(((((..--.(((.........))).))))).))))......... ( -28.60) >DroEre_CAF1 121643 114 - 1 AG-UCAUAAAUUAUGGGCAAUUAAUAUGUAACACGUUAGA--UUCGAAUUCGAUU-GAACAGCCUCUAAUGGCCAAGUCCCU--UGGCCAUUUGUCAGCCUGGAUUUGGCCAUAAAAAAG ..-......((((.((((.........(.....)(((...--.(((....)))..-.))).)))).))))((((((((((..--.(((.........))).))))))))))......... ( -32.00) >DroWil_CAF1 185014 119 - 1 AU-CCAUAAAUAAUGGGCAAUUAAUAUGUAACACGUUAGUCUUUCGAUUUCGAUUUGGGCAACUCUAAAUGGCCAAUUCUUUUCUAGCCAUUUGUCAAUUUUGAUGUGGCCAUAAAAAAC .(-((((.....))))).......((((...(((((((((((.(((....)))...)))).....((((((((.............)))))))).......)))))))..))))...... ( -26.12) >DroYak_CAF1 126186 112 - 1 AG-UCAUAAAUUAUGGGCAAUUAAUAUGUAACACGUUAGA--UUCGAAUUCGAUU-GAACAGCCUCUAAUGGCCAAGUCCUU--UGGCCAUUUGUCAGCCUGGAUUUGGCCAUAAAAA-- ..-......((((.((((.........(.....)(((...--.(((....)))..-.))).)))).))))((((((((((..--.(((.........))).)))))))))).......-- ( -32.70) >DroAna_CAF1 122964 115 - 1 AGGCCAUAAAUUAUGGGCAAUUAAUAUGUAACACGUUAGA--UCCGAAUCCGAUU-CAGAGGCCUCUAAUGGCGGACUCCUC--GGGCCAUUUGUCAGCAAGGAUGUGGCCAUAAAAAAG .(((((((.......(((..((((..((...))..)))).--.((((.((((...-.(((....))).....))))....))--))))).((((....))))..)))))))......... ( -28.90) >consensus AG_UCAUAAAUUAUGGGCAAUUAAUAUGUAACACGUUAGA__UUCGAAUUCGAUU_GAACAGCCUCUAAUGGCCAAGUCCUU__UGGCCAUUUGUCAGCCUGGAUUUGGCCAUAAAAAAG ..............((((..((((..((...))..))))....(((....)))........))))...((((((((((((....((((.....))))....))))))))))))....... (-23.47 = -24.92 + 1.45)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:01 2006