
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,520,379 – 12,520,547 |
| Length | 168 |
| Max. P | 0.999134 |

| Location | 12,520,379 – 12,520,471 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 75.04 |
| Mean single sequence MFE | -28.78 |
| Consensus MFE | -11.02 |
| Energy contribution | -13.22 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.38 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994112 |
| Prediction | RNA |
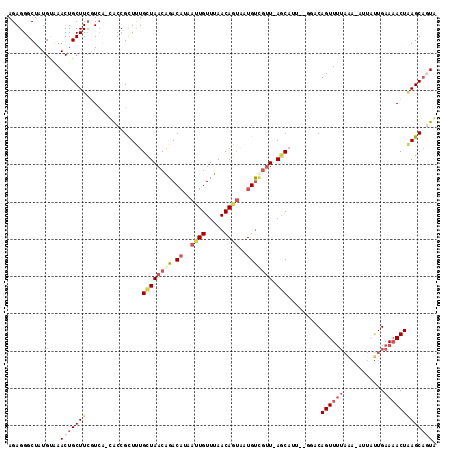
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12520379 92 + 22407834 AGAGGGCUAUGUAAACUGCUUAGUCA-AACCGCUUUGCUAACAGACAAAACUGUUUAACAGUGAUGU---U-AGC-------ACAGUU-----------UGAAAACUAAGCAGUA ..............((((((((((..-....(((.((((((((......((((.....))))..)))---)-)))-------).))).-----------.....)))))))))). ( -31.00) >DroSec_CAF1 92993 112 + 1 AGAGGGCUAUGUAAACUGCUUCGUCG-CACCGUUUUGCUAACAGACAUGAUUGUUUAACAGUAAUGUUGUU-AGCAUU-GGUACAGUUUUAAAAAAUUUUGAACACUAAGCAGUG ..............(((((((.((.(-.((((...(((((((((.(((.((((.....)))).))))))))-)))).)-))).).((((...........)))))).))))))). ( -30.60) >DroSim_CAF1 96345 109 + 1 AGAGGGCUAUGUAAACUGCUUCGUCG-CACCGUUUUGCUAACAGACAUGAUUGUUUAACAGUAAUGUUGUU-AGCUGU---GACAGUUUUAAA-AUUAUUGAAAACUAAGCAGUG ..............(((((((.((((-((.......((((((((.(((.((((.....)))).))))))))-))))))---)))((((((...-.......))))))))))))). ( -30.81) >DroEre_CAF1 90841 114 + 1 AGAGGGCUAUGUAAACUGCUUCAUAACUGCCGCUCUGCUAACAGACAGAAUUGUUUAACAGUCAUGUCGUUUAACAUUUCGGACAGUUUUACA-CUCAUUGAAAACUGAGCACUA (((((((((((..........))))...))).))))(((....((((..((((.....))))..))))...............(((((((.((-.....)))))))))))).... ( -25.90) >DroYak_CAF1 94644 108 + 1 AGAGGGCUAUGCGAACUGCUUCAUCA-UGCCGCUCUGUUAACGGAC-----UGUUUAACAGAAAUGCCGUUUAACAUUAUGCACAGUUUUAGA-CUUACUGAAAACUUAGCUCUA ..((((((((((((.(.((.......-.)).).))(((((((((.(-----((.....))).....))).))))))....)))((((......-...))))......))))))). ( -25.60) >consensus AGAGGGCUAUGUAAACUGCUUCGUCA_CACCGCUUUGCUAACAGACAUAAUUGUUUAACAGUAAUGUCGUU_AGCAUU__GGACAGUUUUAAA_AUUAUUGAAAACUAAGCAGUA ..............(((((((...............((((((((.((..((((.....))))..))))))).))).........((((((...........))))))))))))). (-11.02 = -13.22 + 2.20)

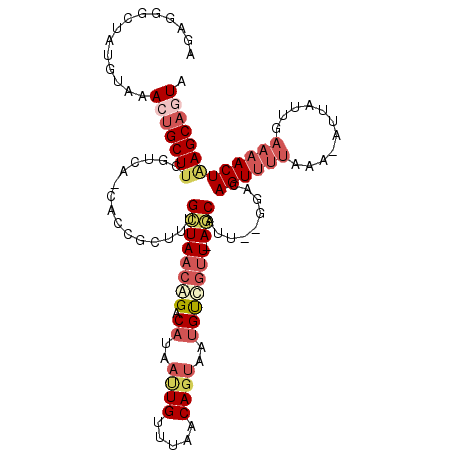
| Location | 12,520,379 – 12,520,471 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 75.04 |
| Mean single sequence MFE | -27.31 |
| Consensus MFE | -8.96 |
| Energy contribution | -7.88 |
| Covariance contribution | -1.08 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.33 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12520379 92 - 22407834 UACUGCUUAGUUUUCA-----------AACUGU-------GCU-A---ACAUCACUGUUAAACAGUUUUGUCUGUUAGCAAAGCGGUU-UGACUAAGCAGUUUACAUAGCCCUCU .(((((((((...(((-----------((((((-------(((-(---(((..((((.....))))......)))))))...))))))-)))))))))))).............. ( -32.10) >DroSec_CAF1 92993 112 - 1 CACUGCUUAGUGUUCAAAAUUUUUUAAAACUGUACC-AAUGCU-AACAACAUUACUGUUAAACAAUCAUGUCUGUUAGCAAAACGGUG-CGACGAAGCAGUUUACAUAGCCCUCU .((((((((((.................)))(((((-..((((-((((((((.(.((.....)).).)))).))))))))....))))-)....))))))).............. ( -26.33) >DroSim_CAF1 96345 109 - 1 CACUGCUUAGUUUUCAAUAAU-UUUAAAACUGUC---ACAGCU-AACAACAUUACUGUUAAACAAUCAUGUCUGUUAGCAAAACGGUG-CGACGAAGCAGUUUACAUAGCCCUCU .(((((((((((((.((....-)).))))))(((---.(((((-((((((((.(.((.....)).).)))).))))))).......))-.))).))))))).............. ( -24.01) >DroEre_CAF1 90841 114 - 1 UAGUGCUCAGUUUUCAAUGAG-UGUAAAACUGUCCGAAAUGUUAAACGACAUGACUGUUAAACAAUUCUGUCUGUUAGCAGAGCGGCAGUUAUGAAGCAGUUUACAUAGCCCUCU ..(..((((........))))-..)..(((((.(((...((((((..((((.((.((.....)).)).))))..))))))...))))))))........................ ( -27.30) >DroYak_CAF1 94644 108 - 1 UAGAGCUAAGUUUUCAGUAAG-UCUAAAACUGUGCAUAAUGUUAAACGGCAUUUCUGUUAAACA-----GUCCGUUAACAGAGCGGCA-UGAUGAAGCAGUUCGCAUAGCCCUCU ....((..((((((.((....-.))))))))..))....((((((.(((.....(((.....))-----).)))))))))(((.((((-((.((((....))))))).)))))). ( -26.80) >consensus UACUGCUUAGUUUUCAAUAAG_UUUAAAACUGUCC__AAUGCU_AACAACAUUACUGUUAAACAAUCAUGUCUGUUAGCAAAGCGGUA_UGACGAAGCAGUUUACAUAGCCCUCU ....(((..((.............................(((.(((((((....((.....))....))).))))))).((((.((.........)).))))))..)))..... ( -8.96 = -7.88 + -1.08)

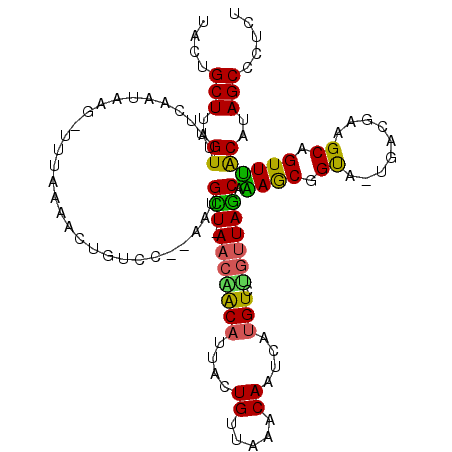

| Location | 12,520,413 – 12,520,511 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.67 |
| Mean single sequence MFE | -27.97 |
| Consensus MFE | -12.62 |
| Energy contribution | -15.62 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.45 |
| SVM decision value | 3.39 |
| SVM RNA-class probability | 0.999134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12520413 98 + 22407834 UGCUAACAGACAAAACUGUUUAACAGUGAUGU---U-AGC-------ACAGUU-----------UGAAAACUAAGCAGUAGUGCUGUAAACAGUAAUGCUGUUUAACAGUUUCGACAGUU ((((((((......((((.....))))..)))---)-)))-------)..(((-----------.((((..(((((((((.(((((....))))).)))))))))....))))))).... ( -32.50) >DroSec_CAF1 93027 118 + 1 UGCUAACAGACAUGAUUGUUUAACAGUAAUGUUGUU-AGCAUU-GGUACAGUUUUAAAAAAUUUUGAACACUAAGCAGUGUUACUGUAAACAGUAAUGCUGUUUAACAGUUUCGACAGUU (((((((((.(((.((((.....)))).))))))))-))))((-((.((.((((...........))))..(((((((((((((((....)))))))))))))))...)).))))..... ( -36.60) >DroSim_CAF1 96379 114 + 1 UGCUAACAGACAUGAUUGUUUAACAGUAAUGUUGUU-AGCUGU---GACAGUUUUAAA-AUUAUUGAAAACUAAGCAGUGAUACUGUAAACAGGAAUGCUGU-UAACAGUUUCGACAGUU .((((((((.(((.((((.....)))).))))))))-)))(((---...((((((...-.......))))))..)))((((.(((((.(((((.....))))-).))))).)).)).... ( -31.20) >DroEre_CAF1 90876 115 + 1 UGCUAACAGACAGAAUUGUUUAACAGUCAUGUCGUUUAACAUUUCGGACAGUUUUACA-CUCAUUGAAAACUGAGCACUACUU----UAACAUAAAUGUUGUUUAACAGACUCAACCGUU ((((....((((..((((.....))))..))))...............(((((((.((-.....)))))))))))))......----(((((....)))))................... ( -18.70) >DroYak_CAF1 94678 113 + 1 UGUUAACGGAC-----UGUUUAACAGAAAUGCCGUUUAACAUUAUGCACAGUUUUAGA-CUUACUGAAAACUUAGCUCUACUU-UCUUAACAUGCAUGUUGUUUAACAGUUUCGACAGUU ((((...((((-----((((.(((((..((((.(((.........((..((((((((.-....))).)))))..)).......-....)))..)))).))))).)))))))).))))... ( -20.85) >consensus UGCUAACAGACAUAAUUGUUUAACAGUAAUGUCGUU_AGCAUU__GGACAGUUUUAAA_AUUAUUGAAAACUAAGCAGUACUACUGUAAACAGGAAUGCUGUUUAACAGUUUCGACAGUU .((((((((.((..((((.....))))..))))))).)))........................(((((..(((((((((.(((((....))))).)))))))))....)))))...... (-12.62 = -15.62 + 3.00)



| Location | 12,520,413 – 12,520,511 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.67 |
| Mean single sequence MFE | -24.02 |
| Consensus MFE | -6.08 |
| Energy contribution | -7.04 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.25 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989419 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12520413 98 - 22407834 AACUGUCGAAACUGUUAAACAGCAUUACUGUUUACAGCACUACUGCUUAGUUUUCA-----------AACUGU-------GCU-A---ACAUCACUGUUAAACAGUUUUGUCUGUUAGCA (((.(.((((((((((.(((((.....(((....))).......((.(((((....-----------))))).-------)).-.---......))))).)))))))))).).))).... ( -25.30) >DroSec_CAF1 93027 118 - 1 AACUGUCGAAACUGUUAAACAGCAUUACUGUUUACAGUAACACUGCUUAGUGUUCAAAAUUUUUUAAAACUGUACC-AAUGCU-AACAACAUUACUGUUAAACAAUCAUGUCUGUUAGCA ..........(((((.((((((.....)))))))))))((((((....))))))......................-..((((-((((((((.(.((.....)).).)))).)))))))) ( -25.60) >DroSim_CAF1 96379 114 - 1 AACUGUCGAAACUGUUA-ACAGCAUUCCUGUUUACAGUAUCACUGCUUAGUUUUCAAUAAU-UUUAAAACUGUC---ACAGCU-AACAACAUUACUGUUAAACAAUCAUGUCUGUUAGCA .......((.(((((.(-((((.....))))).))))).))......(((((((.((....-)).)))))))..---...(((-((((((((.(.((.....)).).)))).))))))). ( -26.30) >DroEre_CAF1 90876 115 - 1 AACGGUUGAGUCUGUUAAACAACAUUUAUGUUA----AAGUAGUGCUCAGUUUUCAAUGAG-UGUAAAACUGUCCGAAAUGUUAAACGACAUGACUGUUAAACAAUUCUGUCUGUUAGCA ((((((((.(((.(((....(((((((......----.(((.(..((((........))))-..)...))).....))))))).)))))).)))))))).((((........)))).... ( -22.10) >DroYak_CAF1 94678 113 - 1 AACUGUCGAAACUGUUAAACAACAUGCAUGUUAAGA-AAGUAGAGCUAAGUUUUCAGUAAG-UCUAAAACUGUGCAUAAUGUUAAACGGCAUUUCUGUUAAACA-----GUCCGUUAACA ....((..(.((((((.((((..((((...((((.(-...((..((..((((((.((....-.))))))))..)).)).).))))...))))...)))).))))-----))...)..)). ( -20.80) >consensus AACUGUCGAAACUGUUAAACAGCAUUACUGUUUACAGAAGUACUGCUUAGUUUUCAAUAAG_UUUAAAACUGUCC__AAUGCU_AACAACAUUACUGUUAAACAAUCAUGUCUGUUAGCA .............(((((...(((....(((((..............(((((((...........)))))))...............((((....)))))))))....)))...))))). ( -6.08 = -7.04 + 0.96)

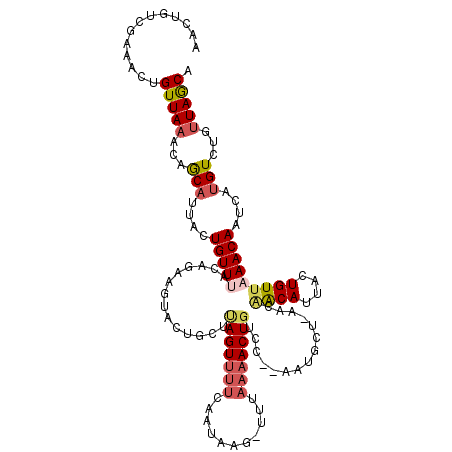
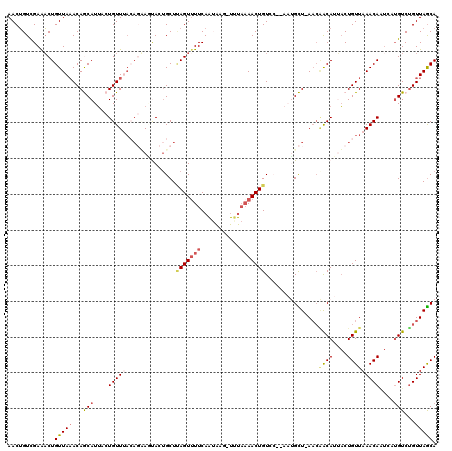
| Location | 12,520,449 – 12,520,547 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 71.77 |
| Mean single sequence MFE | -23.94 |
| Consensus MFE | -9.73 |
| Energy contribution | -11.01 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12520449 98 + 22407834 -------ACAGUU-----------UGAAAACUAAGCAGUAGUGCUGUAAACAGUAAUGCUGUUUAACAGUUUCGACAGUUUUGGAUUGAUAGA-AU-AUGAAGUACACUGCUUGAAUA -------......-----------.......((((((((.(((((.(((((((.....))))))).((..(((..(((((...)))))...))-).-.)).))))))))))))).... ( -25.60) >DroSec_CAF1 93066 115 + 1 AUU-GGUACAGUUUUAAAAAAUUUUGAACACUAAGCAGUGUUACUGUAAACAGUAAUGCUGUUUAACAGUUUCGACAGUUUUGAAUUGAUAGA-GU-AGGAAUCACACUGCUUGAAUU ...-....((((((...(((((((((((.(((((((((((((((((....))))))))))))))...)))))))).))))))))))))...((-((-((........))))))..... ( -31.70) >DroSim_CAF1 96418 113 + 1 UGU---GACAGUUUUAAA-AUUAUUGAAAACUAAGCAGUGAUACUGUAAACAGGAAUGCUGU-UAACAGUUUCGACAGUUUUGAAUUGAUCGAAGUAAGUAAGUACACUGCUUGAAUU ...---...((((((...-.......))))))((((((((..(((((.(((((.....))))-).)))))(((((((((.....)))).)))))...........))))))))..... ( -29.50) >DroEre_CAF1 90916 110 + 1 AUUUCGGACAGUUUUACA-CUCAUUGAAAACUGAGCACUACUU----UAACAUAAAUGUUGUUUAACAGACUCAACCGUUUUA-GCUUAUAGA-AA-AUUAAGCACACUGCUUGAGUA .((((...(((((((.((-.....)))))))))(((.......----(((((....)))))......((((......))))..-)))....))-))-.((((((.....))))))... ( -17.60) >DroYak_CAF1 94713 112 + 1 AUUAUGCACAGUUUUAGA-CUUACUGAAAACUUAGCUCUACUU-UCUUAACAUGCAUGUUGUUUAACAGUUUCGACAGUUUUA-UUUGAU-GA-AU-ACGAAAUACACUGCUUGAGCA ..((((((..(((..(((-....((((....))))........-))).))).)))))).(((((((((((((((...((((..-......-))-))-.))))....)))).))))))) ( -15.30) >consensus AUU__GGACAGUUUUAAA_AUUAUUGAAAACUAAGCAGUACUACUGUAAACAGGAAUGCUGUUUAACAGUUUCGACAGUUUUG_AUUGAUAGA_AU_AGGAAGUACACUGCUUGAAUA ...............................(((((((((.(((((....))))).))))))))).....(((((((((...........................)))).))))).. ( -9.73 = -11.01 + 1.28)

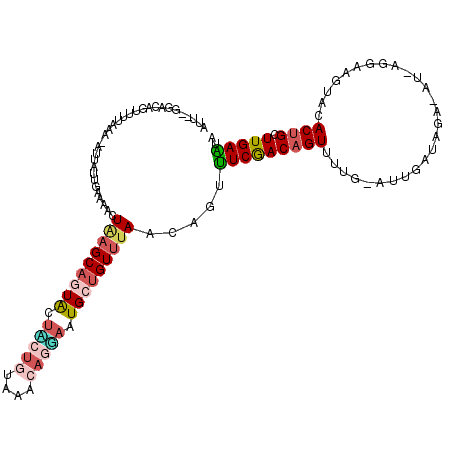
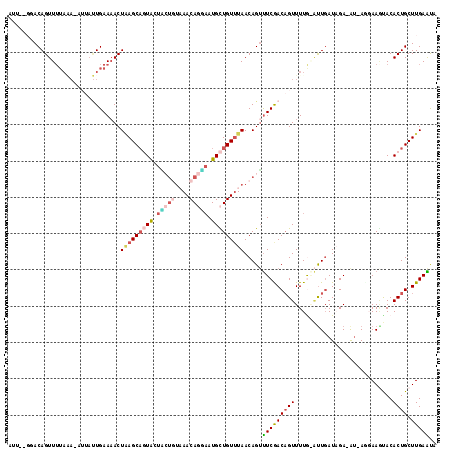
| Location | 12,520,449 – 12,520,547 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 71.77 |
| Mean single sequence MFE | -20.29 |
| Consensus MFE | -7.56 |
| Energy contribution | -8.52 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12520449 98 - 22407834 UAUUCAAGCAGUGUACUUCAU-AU-UCUAUCAAUCCAAAACUGUCGAAACUGUUAAACAGCAUUACUGUUUACAGCACUACUGCUUAGUUUUCA-----------AACUGU------- .....((((((((........-.(-((...((.........))..))).((((.((((((.....))))))))))...))))))))((((....-----------))))..------- ( -17.20) >DroSec_CAF1 93066 115 - 1 AAUUCAAGCAGUGUGAUUCCU-AC-UCUAUCAAUUCAAAACUGUCGAAACUGUUAAACAGCAUUACUGUUUACAGUAACACUGCUUAGUGUUCAAAAUUUUUUAAAACUGUACC-AAU .....((((((((((((....-..-...))).................(((((.((((((.....))))))))))).)))))))))............................-... ( -23.40) >DroSim_CAF1 96418 113 - 1 AAUUCAAGCAGUGUACUUACUUACUUCGAUCAAUUCAAAACUGUCGAAACUGUUA-ACAGCAUUCCUGUUUACAGUAUCACUGCUUAGUUUUCAAUAAU-UUUAAAACUGUC---ACA .....((((((((...........((((((............))))))(((((.(-((((.....))))).)))))..))))))))((((((.((....-)).))))))...---... ( -26.50) >DroEre_CAF1 90916 110 - 1 UACUCAAGCAGUGUGCUUAAU-UU-UCUAUAAGC-UAAAACGGUUGAGUCUGUUAAACAACAUUUAUGUUA----AAGUAGUGCUCAGUUUUCAAUGAG-UGUAAAACUGUCCGAAAU ((((..(((((((((((((..-..-....)))))-...(((((......))))).....))))...)))).----.)))).((..(((((((((.....-)).)))))))..)).... ( -19.20) >DroYak_CAF1 94713 112 - 1 UGCUCAAGCAGUGUAUUUCGU-AU-UC-AUCAAA-UAAAACUGUCGAAACUGUUAAACAACAUGCAUGUUAAGA-AAGUAGAGCUAAGUUUUCAGUAAG-UCUAAAACUGUGCAUAAU .((((.((((((....((((.-..-..-......-.........))))))))))....((((....))))....-.....))))..((((((.((....-.))))))))......... ( -15.15) >consensus UAUUCAAGCAGUGUACUUCAU_AU_UCUAUCAAU_CAAAACUGUCGAAACUGUUAAACAGCAUUACUGUUUACAGAAGUACUGCUUAGUUUUCAAUAAG_UUUAAAACUGUCC__AAU .(((.((((((((.........................(((.(......).))).(((((.....)))))........)))))))))))............................. ( -7.56 = -8.52 + 0.96)

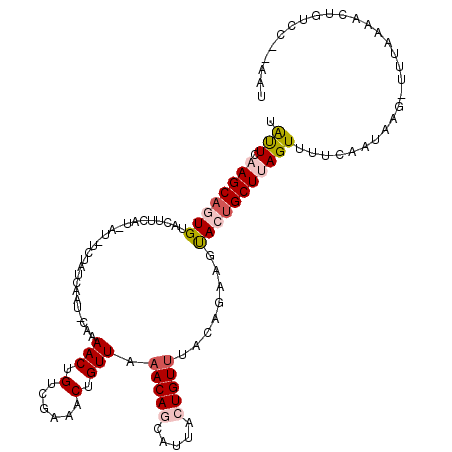
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:55 2006