| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,516,504 – 12,516,599 |
| Length | 95 |
| Max. P | 0.982722 |

| Location | 12,516,504 – 12,516,599 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 77.12 |
| Mean single sequence MFE | -43.87 |
| Consensus MFE | -28.06 |
| Energy contribution | -28.25 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
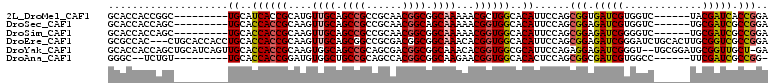
>2L_DroMel_CAF1 12516504 95 + 22407834 GCACCACCGGC---------UGCAUCACCGCAUGUUGCAGCCGCCGCAACGGCGGCAAAAACGCUGGCACAUUCCAGCGGUGAUCGUGGUC------UACGAUCACCGGA ......(((((---------((((.((.....)).)))))))(((((....)))))......(((((......)))))((((((((((...------)))))))))))). ( -48.70) >DroSec_CAF1 89218 95 + 1 GCACCACCAGC---------UGCACCACCGCAAGUUGCAGCCGCCGCAACGGCAGCAAAAACGGUGGCACAUUCCAGCGGAGAUCGUGGUC------UGCGAUCGCCGGA ......((.((---------((..((((((....((((.((((......)))).))))...)))))).......))))((.((((((....------.)))))).)))). ( -39.30) >DroSim_CAF1 92342 95 + 1 GCACCACCAGC---------UGCACCACCGCAAGUUGCAGCCGCCGCAACGGCGGCAAAAACGGUGGCACAUUCCAGCGGAGAUCGGGGUC------UGCGAUCGCCGGA ......((.((---------((..(((((((.....)).(((((((...)))))))......))))).......))))((.(((((.....------..))))).)))). ( -40.40) >DroEre_CAF1 87070 107 + 1 GCGCCAC---CUGCACCACCUGCACCACCGCAAGUUGCAGCGGCCGCGACGGCGGCAAACACGGUGGCACAUUCCAGCGGAGAUCGGGAUCUGCACUUGCGGUCGCCGGA (.(((((---(((((..((.(((......))).))))))...(((((....)))))......)))))).).((((...)))).((((((.((((....)))))).)))). ( -42.80) >DroYak_CAF1 90789 107 + 1 GCACCACCAGCUGCAUCAGUUGCACCACCGCAAGUGGCAGCCGCAGCGACGGCGGCAAACACGGUGGCGCAUUCCAGAGGAGAUCGGGU--UGCGGAUGCGGUUGCU-GA ....((.((((((((((.......(((((((.....)).(((((.......)))))......)))))((((..((.((.....))))..--)))))))))))))).)-). ( -46.60) >DroAna_CAF1 88685 92 + 1 GGGC--UCUGU---------UGCACCACCGGAUGUGGCUGCCGCAGCCACGGCGGCAAGAACGGUGGCACACUCCAGCGGCGAUCGUGGCC------UUCGAUCGCCGG- ((((--.....---------.)).((((((..(...(((((((......)))))))..)..))))))......))..(((((((((.....------..))))))))).- ( -45.40) >consensus GCACCACCAGC_________UGCACCACCGCAAGUUGCAGCCGCCGCAACGGCGGCAAAAACGGUGGCACAUUCCAGCGGAGAUCGGGGUC______UGCGAUCGCCGGA ....................((..((((((....((((.((((......)))).))))...))))))..))......(((.(((((.............))))).))).. (-28.06 = -28.25 + 0.20)

| Location | 12,516,504 – 12,516,599 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 77.12 |
| Mean single sequence MFE | -48.67 |
| Consensus MFE | -35.06 |
| Energy contribution | -36.67 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12516504 95 - 22407834 UCCGGUGAUCGUA------GACCACGAUCACCGCUGGAAUGUGCCAGCGUUUUUGCCGCCGUUGCGGCGGCUGCAACAUGCGGUGAUGCA---------GCCGGUGGUGC ...(((((((((.------....)))))))))(((((......)))))......((((((((((((...(((((.....)))))..))))---------).))))))).. ( -48.60) >DroSec_CAF1 89218 95 - 1 UCCGGCGAUCGCA------GACCACGAUCUCCGCUGGAAUGUGCCACCGUUUUUGCUGCCGUUGCGGCGGCUGCAACUUGCGGUGGUGCA---------GCUGGUGGUGC ....(((((((..------.....))))).((((..(..((..(((((((..((((.(((((....))))).))))...)))))))..))---------.)..)))).)) ( -47.50) >DroSim_CAF1 92342 95 - 1 UCCGGCGAUCGCA------GACCCCGAUCUCCGCUGGAAUGUGCCACCGUUUUUGCCGCCGUUGCGGCGGCUGCAACUUGCGGUGGUGCA---------GCUGGUGGUGC ....(((((((..------.....))))).((((..(..((..(((((((..((((.(((((....))))).))))...)))))))..))---------.)..)))).)) ( -47.10) >DroEre_CAF1 87070 107 - 1 UCCGGCGACCGCAAGUGCAGAUCCCGAUCUCCGCUGGAAUGUGCCACCGUGUUUGCCGCCGUCGCGGCCGCUGCAACUUGCGGUGGUGCAGGUGGUGCAG---GUGGCGC (((((((...((....))((((....)))).)))))))..((((((((.(((..((((((...(((.(((((((.....)))))))))).))))))))))---))))))) ( -57.30) >DroYak_CAF1 90789 107 - 1 UC-AGCAACCGCAUCCGCA--ACCCGAUCUCCUCUGGAAUGCGCCACCGUGUUUGCCGCCGUCGCUGCGGCUGCCACUUGCGGUGGUGCAACUGAUGCAGCUGGUGGUGC ..-.(((.((((..(.(((--.((.((.....)).))..((((((((((((...((.(((((....))))).))....)))))))))))).....))).)...))))))) ( -44.60) >DroAna_CAF1 88685 92 - 1 -CCGGCGAUCGAA------GGCCACGAUCGCCGCUGGAGUGUGCCACCGUUCUUGCCGCCGUGGCUGCGGCAGCCACAUCCGGUGGUGCA---------ACAGA--GCCC -.(((((((((..------.....)))))))))(((...((..((((((.....((.(((((....))))).))......))))))..))---------.))).--.... ( -46.90) >consensus UCCGGCGAUCGCA______GACCACGAUCUCCGCUGGAAUGUGCCACCGUUUUUGCCGCCGUUGCGGCGGCUGCAACUUGCGGUGGUGCA_________GCUGGUGGUGC (((((((((((.............)))))...)))))).(((((((((((..((((.(((((....))))).))))...))))))))))).................... (-35.06 = -36.67 + 1.61)

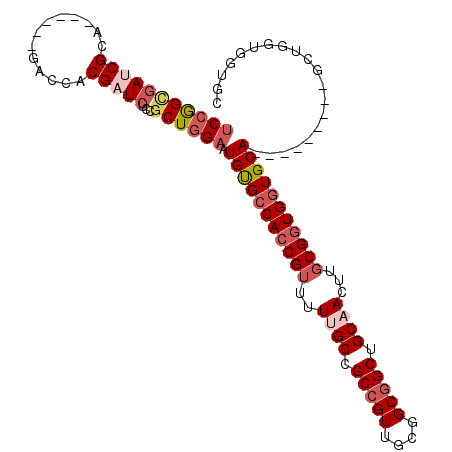
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:50 2006