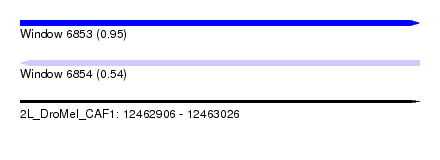
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,462,906 – 12,463,026 |
| Length | 120 |
| Max. P | 0.952983 |

| Location | 12,462,906 – 12,463,026 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.96 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -18.57 |
| Energy contribution | -20.37 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.952983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12462906 120 + 22407834 AACUUUAAGAUAAGCAGCUCUAGCUUAUCAUUGUUAUCAUUGAAUGCGAUAGAUAACUCAAUGUAAAGACGAGGAAGUCGCCAAGAAAAAACGUCUCUCAUUGCAAAACAUUCGGAAAAU ........(((((((.......)))))))...((((((((((....)))).)))))).(((((...(((((......((.....)).....)))))..)))))................. ( -25.40) >DroSec_CAF1 35905 107 + 1 AACUUUAAGAUAAGCAGCUCUAGCUUAUCAUUGUUAUCAUUGAAUGCGAUAGAUAACUCAAUGUAAAGACGAGGAAGUC-------------GUCUCUCUUUGCAGCCCGUUCGGAAAAU ........(((((((.......)))))))...((((((((((....)))).))))))....((((((((.((((.....-------------.))))))))))))..((....))..... ( -28.40) >DroSim_CAF1 37034 107 + 1 AACUUUAAGAUAAGCAGCUCUAGCUUAUCAUUGUUAUCAUUGAAUGCGAUAGAUAACUCAAUGUAAAGACGAGGAAGUC-------------GUCUCUCUUUGCAGCCCGUUCGGAAAAU ........(((((((.......)))))))...((((((((((....)))).))))))....((((((((.((((.....-------------.))))))))))))..((....))..... ( -28.40) >DroEre_CAF1 33004 111 + 1 AACUUUAAGAUAAGCAGGUGUUGCUUA---------UCAUUGAAUGCGAUAGAUUACUCGAUGCAAAGACGAGGAAGUCGGCAAGAAGAAUCACCUAUAUUUGCAAACCAUUUACAAAUU ...((((((((((((((...)))))))---------)).)))))(((((..((((..((..(((...(((......))).))).))..))))........)))))............... ( -23.40) >DroYak_CAF1 35613 106 + 1 AACUUUAAGAUAAGCAGGACUUGCUUAUCAAUGUUAUCAUUGAAUGCGAUAGAUAACUCGAUGCAAAGACGAGG--------------AAUCGUCUAUCUUCGCAAAGCAUUUGGAAAAU ........(((((((((...)))))))))...((((((((((....)))).))))))..(((((...(.(((((--------------(........)))))))...)))))........ ( -26.50) >consensus AACUUUAAGAUAAGCAGCUCUAGCUUAUCAUUGUUAUCAUUGAAUGCGAUAGAUAACUCAAUGUAAAGACGAGGAAGUC_________AA_CGUCUCUCUUUGCAAACCAUUCGGAAAAU ........(((((((.......)))))))...((((((((((....)))).))))))....((((((((.((((...................))))))))))))............... (-18.57 = -20.37 + 1.80)

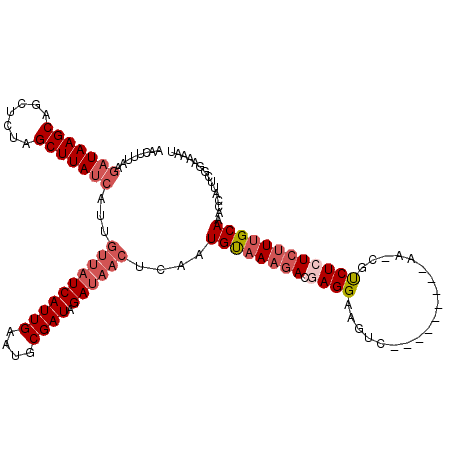
| Location | 12,462,906 – 12,463,026 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.96 |
| Mean single sequence MFE | -25.44 |
| Consensus MFE | -13.27 |
| Energy contribution | -14.55 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12462906 120 - 22407834 AUUUUCCGAAUGUUUUGCAAUGAGAGACGUUUUUUCUUGGCGACUUCCUCGUCUUUACAUUGAGUUAUCUAUCGCAUUCAAUGAUAACAAUGAUAAGCUAGAGCUGCUUAUCUUAAAGUU .................((((((((((((.....((.....))......))))))).))))).((((((.............))))))...(((((((.......)))))))........ ( -25.72) >DroSec_CAF1 35905 107 - 1 AUUUUCCGAACGGGCUGCAAAGAGAGAC-------------GACUUCCUCGUCUUUACAUUGAGUUAUCUAUCGCAUUCAAUGAUAACAAUGAUAAGCUAGAGCUGCUUAUCUUAAAGUU ....(((....)))...(((.(((((((-------------((.....)))))))).).))).((((((.............))))))...(((((((.......)))))))........ ( -24.72) >DroSim_CAF1 37034 107 - 1 AUUUUCCGAACGGGCUGCAAAGAGAGAC-------------GACUUCCUCGUCUUUACAUUGAGUUAUCUAUCGCAUUCAAUGAUAACAAUGAUAAGCUAGAGCUGCUUAUCUUAAAGUU ....(((....)))...(((.(((((((-------------((.....)))))))).).))).((((((.............))))))...(((((((.......)))))))........ ( -24.72) >DroEre_CAF1 33004 111 - 1 AAUUUGUAAAUGGUUUGCAAAUAUAGGUGAUUCUUCUUGCCGACUUCCUCGUCUUUGCAUCGAGUAAUCUAUCGCAUUCAAUGA---------UAAGCAACACCUGCUUAUCUUAAAGUU .((((((((.....))))))))...((((((((((..(((.(((......)))...)))..))).)))).))).........((---------((((((.....))))))))........ ( -25.20) >DroYak_CAF1 35613 106 - 1 AUUUUCCAAAUGCUUUGCGAAGAUAGACGAUU--------------CCUCGUCUUUGCAUCGAGUUAUCUAUCGCAUUCAAUGAUAACAUUGAUAAGCAAGUCCUGCUUAUCUUAAAGUU ...........(((((((((.(..((((((..--------------..))))))...).))).((((((.............))))))...((((((((.....)))))))).)))))). ( -26.82) >consensus AUUUUCCGAAUGGUUUGCAAAGAGAGACG_UU_________GACUUCCUCGUCUUUACAUUGAGUUAUCUAUCGCAUUCAAUGAUAACAAUGAUAAGCUAGAGCUGCUUAUCUUAAAGUU ......................(((((((....................))))))).((((((((..........))))))))........(((((((.......)))))))........ (-13.27 = -14.55 + 1.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:42 2006