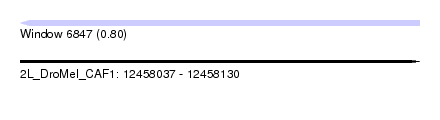
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,458,037 – 12,458,130 |
| Length | 93 |
| Max. P | 0.797569 |

| Location | 12,458,037 – 12,458,130 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.20 |
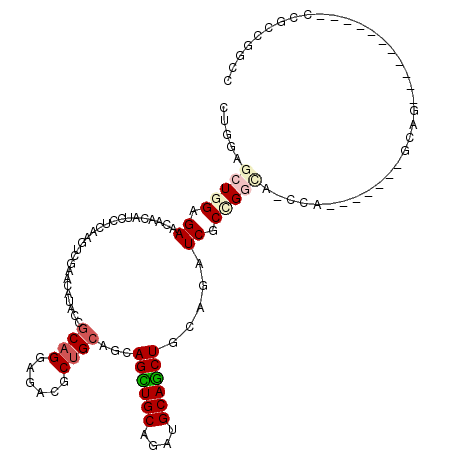
| Mean single sequence MFE | -38.17 |
| Consensus MFE | -17.84 |
| Energy contribution | -18.65 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797569 |
| Prediction | RNA |
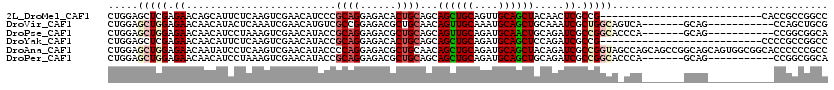
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12458037 93 - 22407834 CUGGAGCUCGAGAACAGCAUUCUCAAGUCGAACAUCCCGCAGGAGACACUGCAGCAGCUGCAGUUGCAGCUACAACUCGCCG---------------------------CACCGCCGGCC ..((((((.(((((.....))))).)))......))).((((......)))).(.((((((....)))))).).....((((---------------------------......)))). ( -29.10) >DroVir_CAF1 36289 102 - 1 CUGGAGCUGGAGAACAACAUACUCAAAUCGAACAUGUCGCCGGAGACGCUGCAACAGUUGCAAAUGCAGCUGCAAAUCGCUGGCAGUCA-------GCAG-----------CCAGCUGCG ..(.((((((...................((...(((.((.(....))).))).(((((((....)))))))....))((((.....))-------))..-----------)))))).). ( -33.80) >DroPse_CAF1 35922 102 - 1 CUGGAGCUGGAGAACAACAUCCUAAAGUCGAACAUACCGCAGGAGACGCUGCAGCAGUUGCAGAUGCAACUGCAGAUCGCCGGCACCCA-------GCAG-----------CCGGCGGCA .((.(((.(((........)))....(((...(........)..)))))).))((((((((....)))))))).(.((((((((.....-------...)-----------)))))))). ( -41.10) >DroYak_CAF1 30564 93 - 1 CUGGAGCUCGAGAACAACAUUCUCAAGUCGAACAUACCGCAGGAGACACUGCAGCAGCUGCAGAUGCAGCUCCAGAUCGCCG---------------------------CCCCGCCGGCC ((((((((.(((((.....)))))..............((((....).((((((...)))))).)))))))))))...((((---------------------------......)))). ( -36.60) >DroAna_CAF1 37314 120 - 1 CUGGAGCUGGAGAACAAUAUCCUCAAGUCGAACAUACCCCAGGAGACGCUGCAACAGCUGCAGAUGCAGCUACAGAUCGCCGGUAGCCAGCAGCCGGCAGCAGUGGCGGCACCCCCCGCC ((((....(((........)))((.....)).......))))....((((((...((((((....)))))).......((((((.(....).)))))).))))))((((......)))). ( -44.90) >DroPer_CAF1 35131 102 - 1 CUGGAGCUGGAGAACAACAUCCUAAAGUCGAACAUACCGCAGGAGACGCUGCAGCAGCUGCAGAUGCAGCUGCAGAUCGCCGGCACCCA-------GCAG-----------CCGGCGGCA .((.(((.(((........)))....(((...(........)..)))))).))((((((((....)))))))).(.((((((((.....-------...)-----------)))))))). ( -43.50) >consensus CUGGAGCUGGAGAACAACAUCCUCAAGUCGAACAUACCGCAGGAGACGCUGCAGCAGCUGCAGAUGCAGCUGCAGAUCGCCGGCA_CCA_______GCAG___________CCGCCGGCC .....(((((.((.........................((((......))))...((((((....)))))).....)).))))).................................... (-17.84 = -18.65 + 0.81)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:35 2006