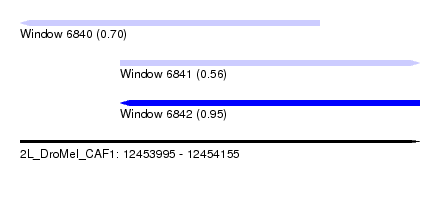
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,453,995 – 12,454,155 |
| Length | 160 |
| Max. P | 0.954444 |

| Location | 12,453,995 – 12,454,115 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -40.73 |
| Consensus MFE | -37.24 |
| Energy contribution | -37.80 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12453995 120 - 22407834 UCAUCGCACCACAGGGGAUCGUGACUAGAAUCGUCCUUGAAGCAAGUCUCUUGCUUUGGUGGUGCAAAUCGAUAUCCUUCGGGCACUUCCGGAUUUAAUUUGAUAAAAGAUCCGGUGGCC .....(((((((.((((((.((.......)).))))))((((((((...)))))))).)))))))....(((......)))(((....((((((((...........))))))))..))) ( -43.00) >DroSec_CAF1 26777 118 - 1 UCGUCGCAC-ACAG-GGAUCGUGACUAGAAUCGUCCUUGAAGCAAGUCUCUUGCUUCGGUGGUGCAAAUCGAUAUCCUUCGGGCACUUCCGGAUUUAUUUUGAUGAAAGAACCGGUGGCC .....((.(-((..-((.((.....((((((.((((..((((((((...))))))))((.(((((...((((......))))))))).))))))..))))))......)).)).))))). ( -36.80) >DroSim_CAF1 26193 120 - 1 UCGUCGCACCACAGGGGAUCGUGACUAGAAUCGUCCUUGAAGCAAGUCUCUUGCUUCGGUGGUGCAAAUCGAUAUCCUUCGGGCACUUCCGGAUUUAUUUUGAUGAAAGAACCGGUGGCC .....(((((((.((((((.((.......)).))))))((((((((...)))))))).)))))))....(((......)))(((....((((.(((.(((....))).)))))))..))) ( -42.40) >consensus UCGUCGCACCACAGGGGAUCGUGACUAGAAUCGUCCUUGAAGCAAGUCUCUUGCUUCGGUGGUGCAAAUCGAUAUCCUUCGGGCACUUCCGGAUUUAUUUUGAUGAAAGAACCGGUGGCC .....(((((((.((((((.((.......)).))))))((((((((...)))))))).)))))))....(((......)))(((....((((.(((.((.....)).))).))))..))) (-37.24 = -37.80 + 0.56)

| Location | 12,454,035 – 12,454,155 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.00 |
| Mean single sequence MFE | -29.97 |
| Consensus MFE | -27.83 |
| Energy contribution | -28.50 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12454035 120 + 22407834 GAAGGAUAUCGAUUUGCACCACCAAAGCAAGAGACUUGCUUCAAGGACGAUUCUAGUCACGAUCCCCUGUGGUGCGAUGAGCAAAUUCUCUCCUCGAAUUUUGAGUGCAAUGGCCUAAUA ..(((((.((((.(((((((((..(((((((...)))))))..(((..((((........)))).)))))))))))).(((......)))...))))))))).((.((....)))).... ( -29.30) >DroSec_CAF1 26817 118 + 1 GAAGGAUAUCGAUUUGCACCACCGAAGCAAGAGACUUGCUUCAAGGACGAUUCUAGUCACGAUCC-CUGU-GUGCGACGAGCAAAUUCUCUCCUCGANNNNNNNNNNNNNNNNNNNNNNN ((.(((.......((((((((..((((((((...))))))))...(((.......))).......-.)).-)))))).(((......))))))))......................... ( -25.30) >DroSim_CAF1 26233 120 + 1 GAAGGAUAUCGAUUUGCACCACCGAAGCAAGAGACUUGCUUCAAGGACGAUUCUAGUCACGAUCCCCUGUGGUGCGACGAGCAAAUUCUCUCCUCGAAUUUUGAGUGCAAUGGCCUAAUA ..(((...(((..(((((((((.((((((((...)))))))).(((..((((........)))).)))))))))))))))((((((((.......))))).....))).....))).... ( -35.30) >consensus GAAGGAUAUCGAUUUGCACCACCGAAGCAAGAGACUUGCUUCAAGGACGAUUCUAGUCACGAUCCCCUGUGGUGCGACGAGCAAAUUCUCUCCUCGAAUUUUGAGUGCAAUGGCCUAAUA ((.(((.......(((((((((.((((((((...))))))))...(((.......)))..........))))))))).(((......))))))))......................... (-27.83 = -28.50 + 0.67)


| Location | 12,454,035 – 12,454,155 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.00 |
| Mean single sequence MFE | -34.40 |
| Consensus MFE | -33.09 |
| Energy contribution | -33.53 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12454035 120 - 22407834 UAUUAGGCCAUUGCACUCAAAAUUCGAGGAGAGAAUUUGCUCAUCGCACCACAGGGGAUCGUGACUAGAAUCGUCCUUGAAGCAAGUCUCUUGCUUUGGUGGUGCAAAUCGAUAUCCUUC ......((....))...........((((((((......)))...(((((((.((((((.((.......)).))))))((((((((...)))))))).))))))).........))))). ( -34.90) >DroSec_CAF1 26817 118 - 1 NNNNNNNNNNNNNNNNNNNNNNNUCGAGGAGAGAAUUUGCUCGUCGCAC-ACAG-GGAUCGUGACUAGAAUCGUCCUUGAAGCAAGUCUCUUGCUUCGGUGGUGCAAAUCGAUAUCCUUC .........................((((((((......)))...((((-((((-((((.((.......)).))))))((((((((...)))))))).)).)))).........))))). ( -30.00) >DroSim_CAF1 26233 120 - 1 UAUUAGGCCAUUGCACUCAAAAUUCGAGGAGAGAAUUUGCUCGUCGCACCACAGGGGAUCGUGACUAGAAUCGUCCUUGAAGCAAGUCUCUUGCUUCGGUGGUGCAAAUCGAUAUCCUUC ....(((..((((............((.(((........))).))(((((((.((((((.((.......)).))))))((((((((...)))))))).)))))))....))))..))).. ( -38.30) >consensus UAUUAGGCCAUUGCACUCAAAAUUCGAGGAGAGAAUUUGCUCGUCGCACCACAGGGGAUCGUGACUAGAAUCGUCCUUGAAGCAAGUCUCUUGCUUCGGUGGUGCAAAUCGAUAUCCUUC .........................((((((((......)))...(((((((.((((((.((.......)).))))))((((((((...)))))))).))))))).........))))). (-33.09 = -33.53 + 0.45)

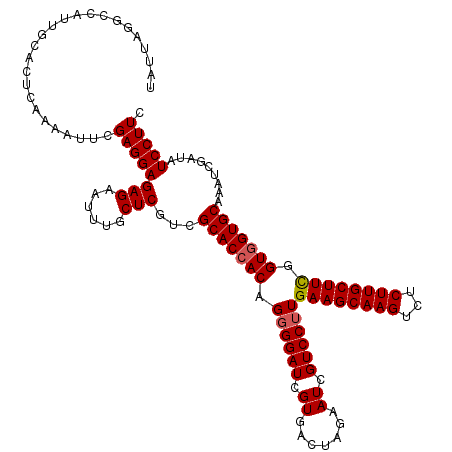
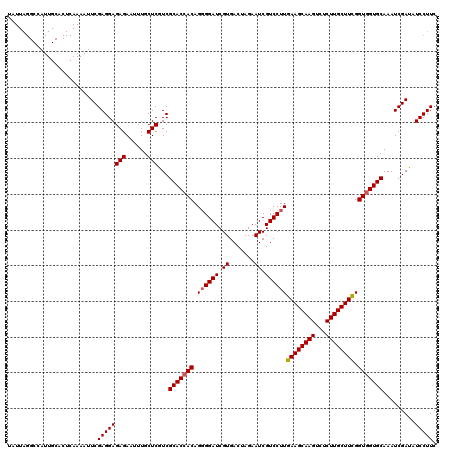
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:30 2006