| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,282,040 – 1,282,144 |
| Length | 104 |
| Max. P | 0.682227 |

| Location | 1,282,040 – 1,282,144 |
|---|---|
| Length | 104 |
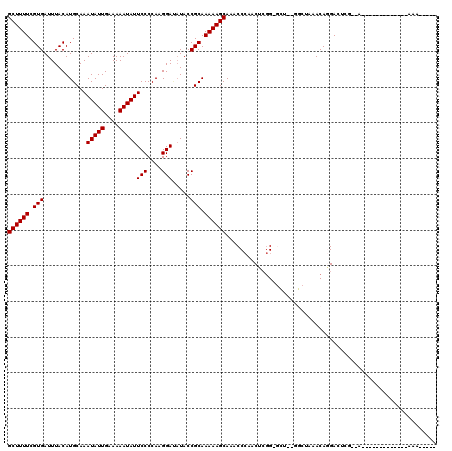
| Sequences | 6 |
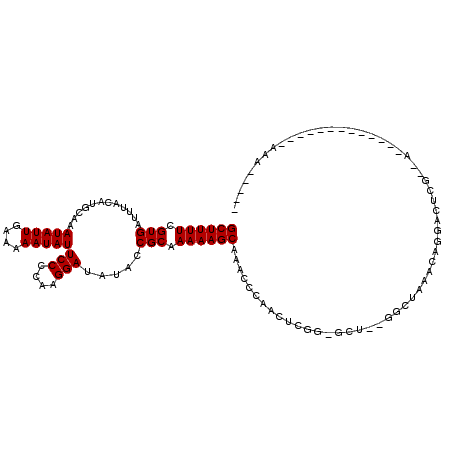
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.20 |
| Mean single sequence MFE | -20.52 |
| Consensus MFE | -12.37 |
| Energy contribution | -12.37 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1282040 104 - 22407834 GCUUUUCGUGAUUUACAUGCAAAUAUUGAAAAAUAUUCCCCAAGGACAUACCGCAAAAAGCAAACCCAGCUCGG-GCUGAGGCUAAACAGGACUCG--A-------------AAAGCAGG ((((((((.(.......(((.((((((....))))))......((.....))))).........((((((....-)))).))..........).))--)-------------)))))... ( -26.20) >DroVir_CAF1 64289 113 - 1 GCUUUUCGUGAUUUACAUGCAAAUAUUGAAAAAUAUUCCCCAAGGA--UACCGCAAAAAGCAAACCAAACU--G-GCUAAAGCUAAACACGAGCGG--UACCAGUAAAAAUAAAAUAAAA ((((((.(((............(((((....)))))(((....)))--...))).)))))).......(((--(-(.((..(((.......)))..--)))))))............... ( -22.80) >DroPse_CAF1 59654 92 - 1 GCUUUUCGUGAUUUACAUGCAAAUAUUGAAAAAUAUUCCCCAAGGAUAUACCGCAAAAAGCAAACCCAACUCGG-A-------AGAACAGGAGAGA--A-------------AAA----- ((((((.(((............(((((....)))))(((....))).....))).))))))....((...((..-.-------.))...)).....--.-------------...----- ( -16.50) >DroYak_CAF1 43268 105 - 1 GCUUUUCGUGAUUUACAUGCAAAUAUUGAAAAAUAUUCCCCAAGGACAUACCGCAAAAAGCAAACCCAACUCGG-GCUGCGGCUAAACAGGACUCGAGG-------------AGAGCAG- (((((((.(((.((...(((.((((((....))))))......((.....)))))....(((..(((.....))-).)))..........)).))).))-------------)))))..- ( -23.70) >DroAna_CAF1 65416 100 - 1 GCUUUUCGUGAUUUACAUGCAAAUAUUGAAAAAUAUUCCCCAAGGAUAUACCGCAAAAAGCAAGCCCCACUCGGUGUUCUGGCUAAACAAGACUCG--C-------------AAA----- ((((((.(((............(((((....)))))(((....))).....))).)))))).((((..((.....))...))))............--.-------------...----- ( -17.40) >DroPer_CAF1 61317 92 - 1 GCUUUUCGUGAUUUACAUGCAAAUAUUGAAAAAUAUUCCCCAAGGAUAUACCGCAAAAAGCAAACCCAACUCGG-A-------AGAACAGGAGAGA--A-------------AAA----- ((((((.(((............(((((....)))))(((....))).....))).))))))....((...((..-.-------.))...)).....--.-------------...----- ( -16.50) >consensus GCUUUUCGUGAUUUACAUGCAAAUAUUGAAAAAUAUUCCCCAAGGAUAUACCGCAAAAAGCAAACCCAACUCGG_GCU__GGCUAAACAGGACUCG__A_____________AAA_____ ((((((.(((............(((((....)))))(((....))).....))).))))))........................................................... (-12.37 = -12.37 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:11 2006