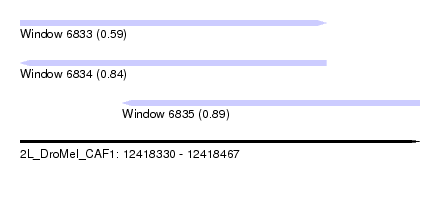
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,418,330 – 12,418,467 |
| Length | 137 |
| Max. P | 0.886697 |

| Location | 12,418,330 – 12,418,435 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 77.13 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -22.42 |
| Energy contribution | -21.78 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
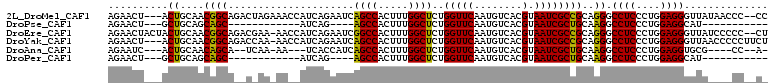
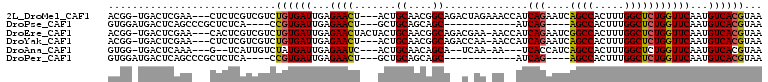
>2L_DroMel_CAF1 12418330 105 + 22407834 GG--GGGUUAUAACCCUCCAGGGAGGCCCUGCGGCGAUUACGUGACAUUGAACCAGAGCCAAAGUGGCUGAUUCUGAUGGUUUCUAGUCUGCCGUUGCAGU---AGUUCU ((--((((....)))).))..((((...((((((((....((.(((...((((((.((((.....))))........))))))...))))).)))))))).---..)))) ( -32.10) >DroPse_CAF1 330723 80 + 1 -----------AUGCCUCCAGGGAGGCCUUGCAGCGAUUACGUGACAUUGAACCAGAGCCAAAGUGGCU----CUGAU------------GCUGCUGCAGC---AGUUCU -----------..(((((....))))).((((((((.....((........))(((((((.....))))----)))..------------..)))))))).---...... ( -32.40) >DroEre_CAF1 240662 107 + 1 AG--GGGGGAUAACCCUCCAGGGAGGCCCUGCGGCGAUUACGUGACAUUGAACCAGAGCCAAAGUGGCCGAUUCUGAUGGUU-UUCGUCUGCCGUUGCAGUAGUAGUUCU .(--(((((....))))))((((..((.((((((((....((.(((...((((((..(((.....))).........)))))-)..))))).))))))))..))..)))) ( -34.90) >DroYak_CAF1 251170 106 + 1 AGAAGGGGGUUAACCCUCCAGGGAGGCCCUGCGGCGAUUACGUGACAUUGAACCAGAGCCAAAGUGGCUGAUUCUGAUGGUU-UUGGUCUGCCGUUGCAGU---AGUUCU ....(((((....)))))(((((...)))))....(((((((..((..(..((((((((((.((.........))..)))))-)))))..)..))..).))---)))).. ( -34.50) >DroAna_CAF1 228704 94 + 1 -U--GG----CGCACCUCCAGGGAGGCCUUGCAGCGAUUACGUGACAUUGAACCAGAGCCAAAGUGGCUGAUGGUGA---UU-UUGA--UGCUGUUGCAGU---GAUUCU -.--.(----((((((((....))))...))).))((((((....((....((((.((((.....))))..))))..---..-.)).--(((....)))))---)))).. ( -28.70) >DroPer_CAF1 333474 80 + 1 -----------AUGCCUCCAGGGAGGCCUUGCAGCGAUUACGUGACAUUGAACCAGAGCCAAAGUGGCU----CUGAU------------GCUGCUGCAGC---AGUUCU -----------..(((((....))))).((((((((.....((........))(((((((.....))))----)))..------------..)))))))).---...... ( -32.40) >consensus _G__GG____UAACCCUCCAGGGAGGCCCUGCAGCGAUUACGUGACAUUGAACCAGAGCCAAAGUGGCUGAUUCUGAU__UU_UU_G__UGCCGUUGCAGU___AGUUCU ..............((((....))))..((((((((.....((........))(((((((.....))))....)))................)))))))).......... (-22.42 = -21.78 + -0.64)


| Location | 12,418,330 – 12,418,435 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 77.13 |
| Mean single sequence MFE | -29.47 |
| Consensus MFE | -18.25 |
| Energy contribution | -17.37 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.839167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12418330 105 - 22407834 AGAACU---ACUGCAACGGCAGACUAGAAACCAUCAGAAUCAGCCACUUUGGCUCUGGUUCAAUGUCACGUAAUCGCCGCAGGGCCUCCCUGGAGGGUUAUAACCC--CC ......---.......((((.(((..((.....)).(((((((((.....)))..))))))...)))..(....)))))(((((...)))))..((((....))))--.. ( -28.30) >DroPse_CAF1 330723 80 - 1 AGAACU---GCUGCAGCAGC------------AUCAG----AGCCACUUUGGCUCUGGUUCAAUGUCACGUAAUCGCUGCAAGGCCUCCCUGGAGGCAU----------- ......---..((((((.(.------------(((((----((((.....))))))))).)........(....)))))))..(((((....)))))..----------- ( -34.30) >DroEre_CAF1 240662 107 - 1 AGAACUACUACUGCAACGGCAGACGAA-AACCAUCAGAAUCGGCCACUUUGGCUCUGGUUCAAUGUCACGUAAUCGCCGCAGGGCCUCCCUGGAGGGUUAUCCCCC--CU ................((((.(((((.-.....)).(((((((((.....))))..)))))...)))..(....)))))(((((...))))).((((.......))--)) ( -28.30) >DroYak_CAF1 251170 106 - 1 AGAACU---ACUGCAACGGCAGACCAA-AACCAUCAGAAUCAGCCACUUUGGCUCUGGUUCAAUGUCACGUAAUCGCCGCAGGGCCUCCCUGGAGGGUUAACCCCCUUCU .(((((---(((((....)))).....-.............((((.....)))).))))))...((..((....))..))((((...))))((((((......)))))). ( -28.30) >DroAna_CAF1 228704 94 - 1 AGAAUC---ACUGCAACAGCA--UCAA-AA---UCACCAUCAGCCACUUUGGCUCUGGUUCAAUGUCACGUAAUCGCUGCAAGGCCUCCCUGGAGGUGCG----CC--A- ......---...((..((((.--....-..---..((((..((((.....)))).))))..........(....)))))....(((((....))))))).----..--.- ( -23.30) >DroPer_CAF1 333474 80 - 1 AGAACU---GCUGCAGCAGC------------AUCAG----AGCCACUUUGGCUCUGGUUCAAUGUCACGUAAUCGCUGCAAGGCCUCCCUGGAGGCAU----------- ......---..((((((.(.------------(((((----((((.....))))))))).)........(....)))))))..(((((....)))))..----------- ( -34.30) >consensus AGAACU___ACUGCAACAGCA__C_AA_AA__AUCAGAAUCAGCCACUUUGGCUCUGGUUCAAUGUCACGUAAUCGCCGCAAGGCCUCCCUGGAGGGUUA____CC__C_ ..........((....((((.....................((((.....))))..(((((........).))))))))..)).((((....)))).............. (-18.25 = -17.37 + -0.89)
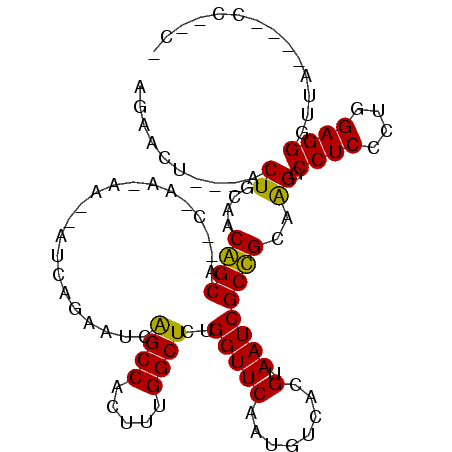

| Location | 12,418,365 – 12,418,467 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 76.75 |
| Mean single sequence MFE | -29.43 |
| Consensus MFE | -16.05 |
| Energy contribution | -15.88 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12418365 102 - 22407834 ACGG-UGACUCGAA---CUCUCGUCGUCUGUGAUUGAGAACU---ACUGCAACGGCAGACUAGAAACCAUCAGAAUCAGCCACUUUGGCUCUGGUUCAAUGUCACGUAA ...(-((((..(((---(..((...((((((..(((((....---.)).)))..))))))..))......((((....(((.....)))))))))))...))))).... ( -28.10) >DroPse_CAF1 330749 86 - 1 GUGGAUGACUCAGCCCGCUCUCA----CCGUGAUUGAGAACU---GCUGCAGCAGC------------AUCAG----AGCCACUUUGGCUCUGGUUCAAUGUCACGUAA ((((..........)))).....----.((((((...(((((---((((...))))------------).(((----((((.....)))))))))))...))))))... ( -32.10) >DroEre_CAF1 240697 104 - 1 ACGG-UGACUCGAA---CACUCGUCGUCUGUGAUUGAGAACUACUACUGCAACGGCAGACGAA-AACCAUCAGAAUCGGCCACUUUGGCUCUGGUUCAAUGUCACGUAA ...(-((((..(((---(.....((((((((..(((((........)).)))..)))))))).-......((((....(((.....)))))))))))...))))).... ( -32.30) >DroYak_CAF1 251207 101 - 1 ACGG-UGACUCGAA---CUCUCGUCGUCUGUGAUUGAGAACU---ACUGCAACGGCAGACCAA-AACCAUCAGAAUCAGCCACUUUGGCUCUGGUUCAAUGUCACGUAA ...(-((((..(((---(.......((((((..(((((....---.)).)))..))))))...-......((((....(((.....)))))))))))...))))).... ( -27.60) >DroAna_CAF1 228734 94 - 1 GUGG-UGACUCAAA---G--UCAUUGUCUAUGAUUGAGAAUC---ACUGCAACAGCA--UCAA-AA---UCACCAUCAGCCACUUUGGCUCUGGUUCAAUGUCACGUAA (..(-(((.((..(---(--((((.....))))))..)).))---))..).......--....-..---..((((..((((.....)))).)))).............. ( -24.40) >DroPer_CAF1 333500 86 - 1 GUGGAUGACUCAGCCCGCUCUCA----CCGUGAUUGAGAACU---GCUGCAGCAGC------------AUCAG----AGCCACUUUGGCUCUGGUUCAAUGUCACGUAA ((((..........)))).....----.((((((...(((((---((((...))))------------).(((----((((.....)))))))))))...))))))... ( -32.10) >consensus ACGG_UGACUCAAA___CUCUCAUCGUCUGUGAUUGAGAACU___ACUGCAACAGCA__C_AA_AA__AUCAGAAUCAGCCACUUUGGCUCUGGUUCAAUGUCACGUAA ............................((((((...((((.......((....))..............(((....((((.....)))))))))))...))))))... (-16.05 = -15.88 + -0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:23 2006