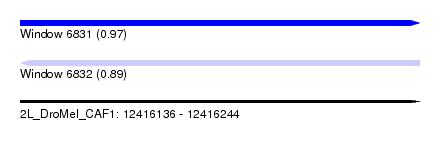
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,416,136 – 12,416,244 |
| Length | 108 |
| Max. P | 0.971287 |

| Location | 12,416,136 – 12,416,244 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.40 |
| Mean single sequence MFE | -28.88 |
| Consensus MFE | -22.18 |
| Energy contribution | -22.18 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971287 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
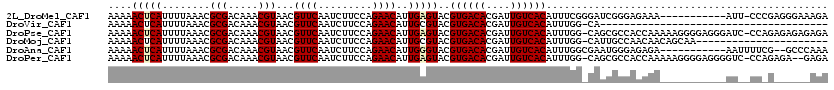
>2L_DroMel_CAF1 12416136 108 + 22407834 UCUUUCCCUCGGG-AAU-----------UUUCUCCCGAUCCCGAAAUGUGACAAUCGUGUCACGUACUCAAUGUUCUGGAAGAUUGAACGUUACGUUUGUCGCGUUUAAAAUGAGUUUUU ........(((((-(..-----------....)))))).....(((((((((((.((((..((((..(((((.((....)).))))))))))))).)))))))))))............. ( -33.00) >DroVir_CAF1 315272 81 + 1 --------------------------------------UG-CCAAAUGUGACAAUCGUGUCACGUACGCAAUGUUCUGGAAGAUUGAACGUUACGUUUGUCGCGUUUAAAAUGAGUUUUU --------------------------------------..-..(((((((((((.((((..((((...((((.((....)).)))).)))))))).)))))))))))............. ( -22.90) >DroPse_CAF1 328634 118 + 1 UCUCUCUCUCUGG-GAUCCCUCCCCUUUUUGGUGGCGCUG-CCAAAUGUGACAAUCGUGUCACGUACUCAAUGUUCUGGAAGAUUGAACGUUACGUUUGUCGCGUUUAAAAUGAGUUUUU .......(((.((-((....))))......(((......)-))(((((((((((.((((..((((..(((((.((....)).))))))))))))).))))))))))).....)))..... ( -32.50) >DroMoj_CAF1 321259 97 + 1 ----------------------UUGCUGUUGUUGGCAAUG-CCAAAUGUGACAAUCGUGUCACGUACGCAAUGUUCUGGAAGAUUGAACGUUACGUUUGUCGCGUUUAAAAUGAGUUUUU ----------------------(((((......)))))..-..(((((((((((.((((..((((...((((.((....)).)))).)))))))).)))))))))))............. ( -26.80) >DroAna_CAF1 226735 107 + 1 UUUGGGC--CGAAAAUU-----------UCUCUCCCAUUCGCCAAAUGUGACAAUCGUGUCACGUACCCAAUGUUCUGGAAGAUUGAACGUUACGUUUGUCGCGUUUAAAAUGAGUUUUU ...(((.--.((.....-----------..)).)))(((((..(((((((((((.((((..((((...((((.((....)).)))).)))))))).)))))))))))....))))).... ( -25.70) >DroPer_CAF1 331355 116 + 1 UCUC--UCUCUGG-GACCCCUCCCCUUUUUGGUGGCGCUG-CCAAAUGUGACAAUCGUGUCACGUACUCAAUGUUCUGGAAGAUUGAACGUUACGUUUGUCGCGUUUAAAAUGAGUUUUU ....--.(((.((-((....))))......(((......)-))(((((((((((.((((..((((..(((((.((....)).))))))))))))).))))))))))).....)))..... ( -32.40) >consensus UCU______C_GG__AU___________UUGCUGGCGAUG_CCAAAUGUGACAAUCGUGUCACGUACUCAAUGUUCUGGAAGAUUGAACGUUACGUUUGUCGCGUUUAAAAUGAGUUUUU ...........................................(((((((((((.((((..((((...((((.((....)).)))).)))))))).)))))))))))............. (-22.18 = -22.18 + 0.00)

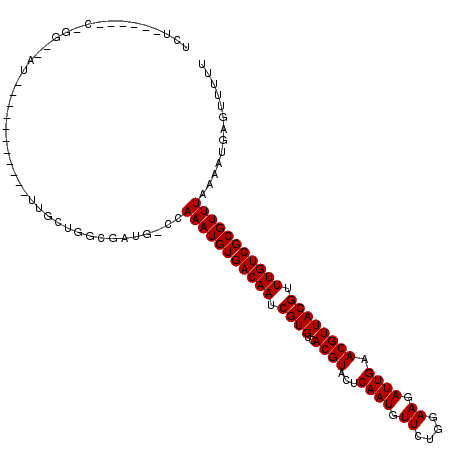
| Location | 12,416,136 – 12,416,244 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.40 |
| Mean single sequence MFE | -25.07 |
| Consensus MFE | -14.20 |
| Energy contribution | -14.45 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.885106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12416136 108 - 22407834 AAAAACUCAUUUUAAACGCGACAAACGUAACGUUCAAUCUUCCAGAACAUUGAGUACGUGACACGAUUGUCACAUUUCGGGAUCGGGAGAAA-----------AUU-CCCGAGGGAAAGA ....(((((........(((.....)))...((((.........))))..)))))..((((((....)))))).((((((((((....))..-----------..)-)))))))...... ( -27.50) >DroVir_CAF1 315272 81 - 1 AAAAACUCAUUUUAAACGCGACAAACGUAACGUUCAAUCUUCCAGAACAUUGCGUACGUGACACGAUUGUCACAUUUGG-CA-------------------------------------- .................((.....((((((.((((.........)))).))))))..((((((....)))))).....)-).-------------------------------------- ( -19.10) >DroPse_CAF1 328634 118 - 1 AAAAACUCAUUUUAAACGCGACAAACGUAACGUUCAAUCUUCCAGAACAUUGAGUACGUGACACGAUUGUCACAUUUGG-CAGCGCCACCAAAAAGGGGAGGGAUC-CCAGAGAGAGAGA .....(((.((((....(.(((((.(((.(((((((((.((....)).)))))..))))...))).))))).)...(((-(...))))........((((....))-)).)))))))... ( -27.40) >DroMoj_CAF1 321259 97 - 1 AAAAACUCAUUUUAAACGCGACAAACGUAACGUUCAAUCUUCCAGAACAUUGCGUACGUGACACGAUUGUCACAUUUGG-CAUUGCCAACAACAGCAA---------------------- .................((.....((((((.((((.........)))).))))))..((((((....))))))..((((-(...))))).....))..---------------------- ( -23.10) >DroAna_CAF1 226735 107 - 1 AAAAACUCAUUUUAAACGCGACAAACGUAACGUUCAAUCUUCCAGAACAUUGGGUACGUGACACGAUUGUCACAUUUGGCGAAUGGGAGAGA-----------AAUUUUCG--GCCCAAA ................(((.....((.(((.((((.........)))).))).))..((((((....)))))).....)))..((((.((((-----------...)))).--.)))).. ( -24.20) >DroPer_CAF1 331355 116 - 1 AAAAACUCAUUUUAAACGCGACAAACGUAACGUUCAAUCUUCCAGAACAUUGAGUACGUGACACGAUUGUCACAUUUGG-CAGCGCCACCAAAAAGGGGAGGGGUC-CCAGAGA--GAGA .....(((.........(.(((((.(((.(((((((((.((....)).)))))..))))...))).))))).).(((((-..((.((.((......))..)).)).-)))))))--)... ( -29.10) >consensus AAAAACUCAUUUUAAACGCGACAAACGUAACGUUCAAUCUUCCAGAACAUUGAGUACGUGACACGAUUGUCACAUUUGG_CAGCGCCACCAA___________AU__CC_G______AGA ....(((((........(((.....)))...((((.........))))..)))))..((((((....))))))............................................... (-14.20 = -14.45 + 0.25)
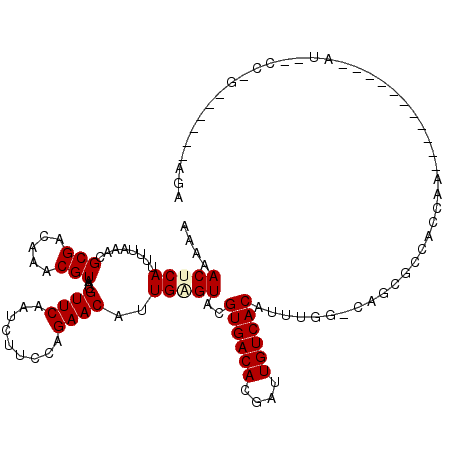
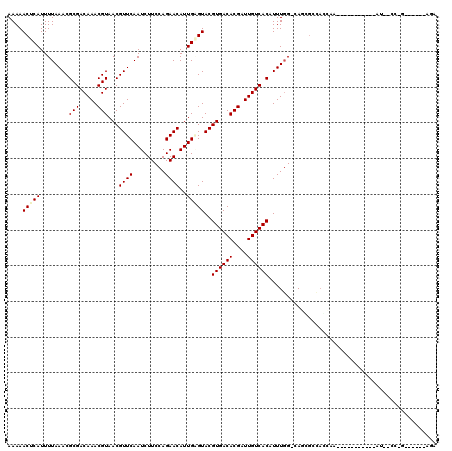
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:21 2006