| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,412,384 – 12,412,498 |
| Length | 114 |
| Max. P | 0.500000 |

| Location | 12,412,384 – 12,412,498 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.70 |
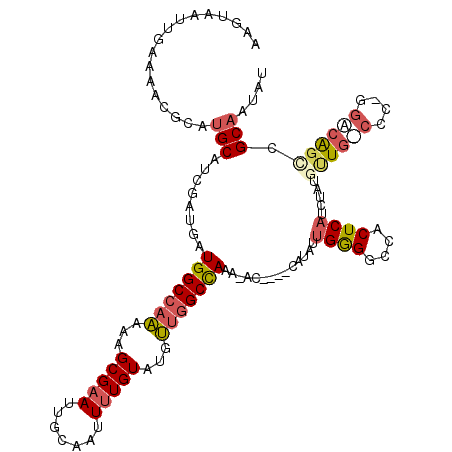
| Mean single sequence MFE | -28.15 |
| Consensus MFE | -19.63 |
| Energy contribution | -20.78 |
| Covariance contribution | 1.15 |
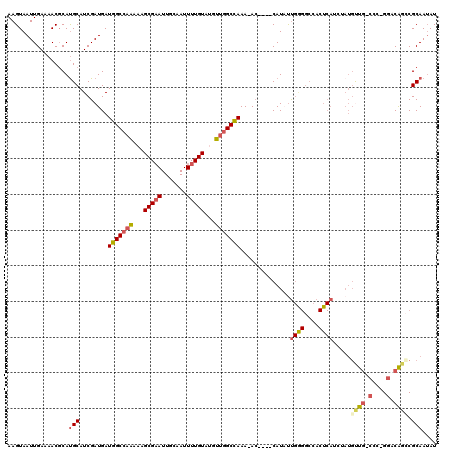
| Combinations/Pair | 1.32 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
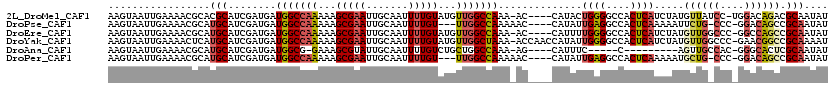
>2L_DroMel_CAF1 12412384 114 - 22407834 AAGUAAUUGAAAACGCACGCAUCGAUGAUGGCCAAAAAGCGAAUUGCAAUUUUGUAUGUUGGCCAAA-AC----CAUACUGGGGCCACUCAUCUAUGUUAUCC-UGGACAGACGCAAUAU ..................((.((((((((((((.....((.(((((((....)))).))).))....-.(----(.....))))))).)))))..((((....-..)))))).))..... ( -27.50) >DroPse_CAF1 324671 111 - 1 AAGUAAUUGAAAACGCAUGCAUCGAUGAUGGCCAAAAAGCGAAUUGCAAUUUUGU---UUGGCCAAAAAC----CAUAUUGAGGCCACUCAAAAAUUCUG-CCC-GGACAGCCGCAAUAU ..............((..(((.......(((((((...(((((.......)))))---))))))).....----....(((((....)))))......))-)..-((....))))..... ( -23.40) >DroEre_CAF1 234922 114 - 1 AAGUAAUUGAAAACGCAUGCAUCGAUGAUGGCCAAAAAGCGAAUUGCAAUUUUGUAUGUUGGCCAAA-AC----CAUUUUGGGGCCACUCAUCUAUGUUGGCCC-GGCCAGCCGCAAUAU .................(((...((((((((((.....((.(((((((....)))).))).))....-.(----(.....))))))).)))))...((((((..-.)))))).))).... ( -32.60) >DroYak_CAF1 245122 118 - 1 AAGUAAUUGAAAACUCAUGCAUCGAUGAUGGCCAAAAAGCGAAUUGCAAUUUUGUAUGUUGGCUAAA-ACCAACCAUAUUGGGGCCACUCAUCUAUGUUGGCCC-GAACGGCCGCAAAAU .................(((...((((((((((....(((.(((((((....)))).))).)))...-.((((.....))))))))).)))))......((((.-....))))))).... ( -35.50) >DroAna_CAF1 223431 99 - 1 AAGUAAUUGAAAACGCAUGCAUCGAUGAUGGCG-GAAAGCGUAUUGCAAUUUUGUCUGCUGGCCAAA-AG----CAUUUC-----C---------AGUUGCCAC-GGGCACUCGCAAUAU .............(((...((....))...)))-......(((((((.....((((((.((((....-((----(.....-----.---------.))))))))-)))))...))))))) ( -22.00) >DroPer_CAF1 327324 111 - 1 AAGUAAUUGAAAACGCAUGCAUCGAUGAUGGCCAAAAAGCGAAUUGCAAUUUUGU---UUGGCCAAAAAC----CAUAUUGAGGCCACUCAAAAAUGCUG-CCC-GGACAGCCGCAAUAU .....((((.....(((.((((......(((((((...(((((.......)))))---))))))).....----....(((((....)))))..))))))-).(-((....))))))).. ( -27.90) >consensus AAGUAAUUGAAAACGCAUGCAUCGAUGAUGGCCAAAAAGCGAAUUGCAAUUUUGUAUGUUGGCCAAA_AC____CAUAUUGGGGCCACUCAUCUAUGUUG_CCC_GGACAGCCGCAAUAU .................(((........(((((((...(((((.......)))))...)))))))..............((((....)))).....((((((....)))))).))).... (-19.63 = -20.78 + 1.15)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:19 2006