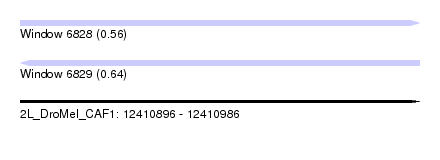
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,410,896 – 12,410,986 |
| Length | 90 |
| Max. P | 0.639211 |

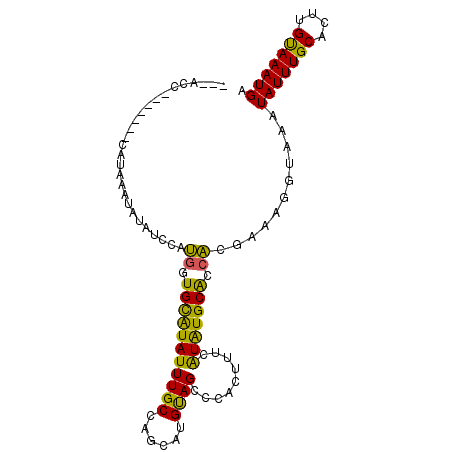
| Location | 12,410,896 – 12,410,986 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 79.65 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -11.22 |
| Energy contribution | -11.67 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12410896 90 + 22407834 UCAUUUACAAGUGCAAAUAUUUACCUUUGGUGUUGCAUAUGAAAGUGGGCUACCUGCUGGCAAAUAUGCACCAUGGAUAUAUUUGUU-------GGU--- ......((....((((((((...((..((((((...((.((..((..((...))..))..)).))..)))))).))..)))))))).-------.))--- ( -24.00) >DroVir_CAF1 308053 90 + 1 UCAUUUACAAGUGCAAAUAUUUACCUUUUGCGGCGCAUACGAAAGUGGGCUACAGCCUGGCAAAUAUGCAGCGUGGAAAUUUUAAUG-------CAU--- ..........(((((...((((.((...((((((((.(((....))).))....)))..(((....))).))).)))))).....))-------)))--- ( -21.30) >DroPse_CAF1 322715 100 + 1 UCAUUUACAAGUGCAAAUAUUUACAUUUCGUGGUGCAUAUGAAAGUGGGCUACAUGCUGGCAAAUAUGCACCAUGGAUAUAUUUAUGGAAUGGUAGUACU .........(((((...(((((.(((((((((((((((((....((.(((.....))).))..)))))))))))))).......))))))))...))))) ( -32.11) >DroMoj_CAF1 314003 90 + 1 UCAUUUACAAGUGCAAAUAUUUACCUUUUAUGGCGCACAUGAAAGUGGGCUACAGCCGGGCAAAUAUGCAGCAUGGAUGUUUAUGUG-------CAU--- .(((..(((.((((..((((((.(((....((((.(((......))).)))).....))).))))))...))))...)))..)))..-------...--- ( -23.00) >DroAna_CAF1 221822 90 + 1 UCAUUUGCAAGUGCAAAUAUUUACAAUUCGUGGUGCAUACGAAAGUGGGCUGCCCUUCGGCAAAUACACACUAUGGAUAUAUUUAUU-------GGC--- ..((((((....)))))).......(((((((((((.(((....))).))((((....)))).......))))))))).........-------...--- ( -24.30) >DroPer_CAF1 325404 100 + 1 UCAUUUACAAGUGCAAAUAUUUACAUUUCGUGGUGCAUAUGAAAGUGGGCUACAUGCUGGCAAAUAUGCACCAUGGAUAUAUUUAUGGAAAGGUAGUACU .........(((((............((((((((((((((....((.(((.....))).))..))))))))))))))..(((((......)))))))))) ( -31.20) >consensus UCAUUUACAAGUGCAAAUAUUUACAUUUCGUGGUGCAUAUGAAAGUGGGCUACAUGCUGGCAAAUAUGCACCAUGGAUAUAUUUAUG_______AGU___ ..........(((((.....((((.(((((((.....)))))))))))((((.....)))).....)))))............................. (-11.22 = -11.67 + 0.45)

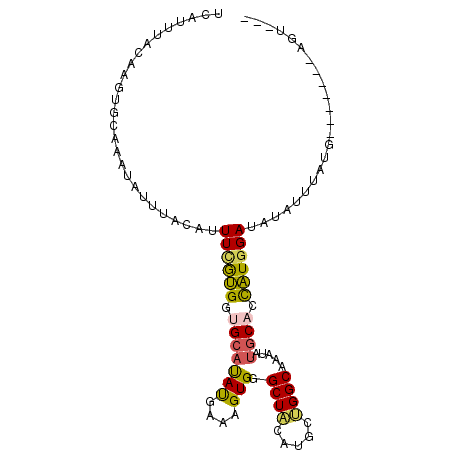
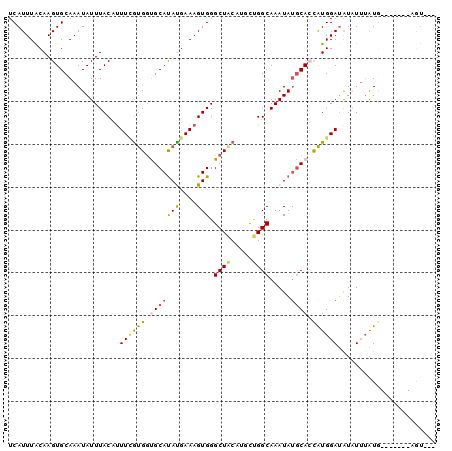
| Location | 12,410,896 – 12,410,986 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 79.65 |
| Mean single sequence MFE | -19.21 |
| Consensus MFE | -11.99 |
| Energy contribution | -10.75 |
| Covariance contribution | -1.24 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12410896 90 - 22407834 ---ACC-------AACAAAUAUAUCCAUGGUGCAUAUUUGCCAGCAGGUAGCCCACUUUCAUAUGCAACACCAAAGGUAAAUAUUUGCACUUGUAAAUGA ---...-------........((((..(((((((((((((((....))))).........))))))...))))..))))..(((((((....))))))). ( -17.50) >DroVir_CAF1 308053 90 - 1 ---AUG-------CAUUAAAAUUUCCACGCUGCAUAUUUGCCAGGCUGUAGCCCACUUUCGUAUGCGCCGCAAAAGGUAAAUAUUUGCACUUGUAAAUGA ---...-------.......((((.((.(.((((((((((((.(((....)))..........(((...)))...))))))))..))))).)).)))).. ( -18.00) >DroPse_CAF1 322715 100 - 1 AGUACUACCAUUCCAUAAAUAUAUCCAUGGUGCAUAUUUGCCAGCAUGUAGCCCACUUUCAUAUGCACCACGAAAUGUAAAUAUUUGCACUUGUAAAUGA ..................((((.((..((((((((((......((.....))........)))))))))).)).))))...(((((((....))))))). ( -19.34) >DroMoj_CAF1 314003 90 - 1 ---AUG-------CACAUAAACAUCCAUGCUGCAUAUUUGCCCGGCUGUAGCCCACUUUCAUGUGCGCCAUAAAAGGUAAAUAUUUGCACUUGUAAAUGA ---..(-------(((((.............(((....)))..(((....))).......))))))(((......)))...(((((((....))))))). ( -20.80) >DroAna_CAF1 221822 90 - 1 ---GCC-------AAUAAAUAUAUCCAUAGUGUGUAUUUGCCGAAGGGCAGCCCACUUUCGUAUGCACCACGAAUUGUAAAUAUUUGCACUUGCAAAUGA ---..(-------(((............((((.((...((((....)))))).)))).((((.......))))))))....(((((((....))))))). ( -20.30) >DroPer_CAF1 325404 100 - 1 AGUACUACCUUUCCAUAAAUAUAUCCAUGGUGCAUAUUUGCCAGCAUGUAGCCCACUUUCAUAUGCACCACGAAAUGUAAAUAUUUGCACUUGUAAAUGA ..................((((.((..((((((((((......((.....))........)))))))))).)).))))...(((((((....))))))). ( -19.34) >consensus ___ACC_______CAUAAAUAUAUCCAUGGUGCAUAUUUGCCAGCAUGUAGCCCACUUUCAUAUGCACCACGAAAGGUAAAUAUUUGCACUUGUAAAUGA ...........................((.(((((((((((......)))).........))))))).))...........(((((((....))))))). (-11.99 = -10.75 + -1.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:18 2006