
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,403,021 – 12,403,118 |
| Length | 97 |
| Max. P | 0.913052 |

| Location | 12,403,021 – 12,403,118 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.02 |
| Mean single sequence MFE | -22.67 |
| Consensus MFE | -13.90 |
| Energy contribution | -13.52 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
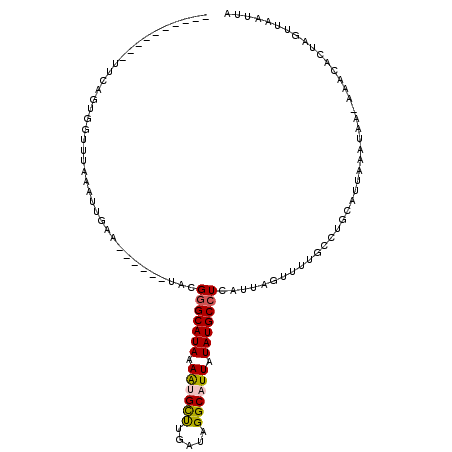
>2L_DroMel_CAF1 12403021 97 + 22407834 UAAUUAACUAGUGUUUUUUAUUUAAUGCAGGCAAAACUAAUGAGGCAUAUAAUGCCUAUCAAGCAUUUUAUGCCCGUA------UUCAAUUUAAACCACUGAA---------- ........(((((.(((..(((.(((((((......))...(.((((((.(((((.......))))).))))))))))------)).)))..))).)))))..---------- ( -23.70) >DroSec_CAF1 226808 95 + 1 UAAUUACCUAGUGUUU-UUAUUUAAUGCAGGCAAAACUAAUGAGGCAUAUAAUGCCUAUCAAGCAUUUUAUGCCCGUA------UUCAAUUUAAAACAGUGA----------- ........((.(((((-(.(((.(((((((......))...(.((((((.(((((.......))))).))))))))))------)).)))..)))))).)).----------- ( -22.40) >DroSim_CAF1 234393 96 + 1 UAAUUAACUAGUGUUUUUUAUUUAAUGCAGGCAAAACUAAUGAGGCAUAUAAUGCCUAUCAAGCAUUUUAUGCCCGUA------UUCAAUUUAAAACAGUGA----------- ........((.((((((..(((.(((((((......))...(.((((((.(((((.......))))).))))))))))------)).)))..)))))).)).----------- ( -24.00) >DroEre_CAF1 225674 105 + 1 UAAUUAACUAGUGCUU-UUAUUUAAUGCAGGGAAAACUAAUGAGGCAUAUAAUGCCUAUCAGGCGUUUUAUGCCCGUA------CUCAAUUUAAACCACUGAAAUCUCCAGC- ........(((((...-...........((......)).(((.((((((.((((((.....)))))).))))))))).------............)))))...........- ( -22.50) >DroYak_CAF1 235645 111 + 1 UAAUUAACUAGUGUUU-UUAUUUAAUGCAGUGAAAACUAAUGAGGCAUAUAAUGCCUAUCAAGCAUUUUAUGCCCGUAUUCGUAUUCAAUUUAAACCACUGAAAUCUCCACC- ........((((((((-..(((.((((((((....))).(((.((((((.(((((.......))))).)))))))))....))))).)))..))).)))))...........- ( -27.30) >DroAna_CAF1 213410 105 + 1 UAAUUAACUA-UGCUG-UUUUUUAAUGCACAAAGAACUAAUGAGGCAUACAAUGCCCAUGGAACGCUUUAUGCUCAUA------AUCGAUUUAAAUCAAGCCUAUCUCUAUCG ..........-.(((.-...(((((.(((.((((.....(((.(((((...))))))))......)))).)))((...------...)).)))))...)))............ ( -16.10) >consensus UAAUUAACUAGUGUUU_UUAUUUAAUGCAGGCAAAACUAAUGAGGCAUAUAAUGCCUAUCAAGCAUUUUAUGCCCGUA______UUCAAUUUAAACCACUGAA__________ ...........(((............)))..........(((.((((((.(((((.......))))).))))))))).................................... (-13.90 = -13.52 + -0.39)


| Location | 12,403,021 – 12,403,118 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.02 |
| Mean single sequence MFE | -23.07 |
| Consensus MFE | -14.13 |
| Energy contribution | -14.22 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12403021 97 - 22407834 ----------UUCAGUGGUUUAAAUUGAA------UACGGGCAUAAAAUGCUUGAUAGGCAUUAUAUGCCUCAUUAGUUUUGCCUGCAUUAAAUAAAAAACACUAGUUAAUUA ----------....(((((..(((((((.------...(((((((.((((((.....)))))).)))))))..))))))).))).)).......................... ( -23.00) >DroSec_CAF1 226808 95 - 1 -----------UCACUGUUUUAAAUUGAA------UACGGGCAUAAAAUGCUUGAUAGGCAUUAUAUGCCUCAUUAGUUUUGCCUGCAUUAAAUAA-AAACACUAGGUAAUUA -----------..........(((((((.------...(((((((.((((((.....)))))).)))))))..)))))))((((((..........-......)))))).... ( -21.29) >DroSim_CAF1 234393 96 - 1 -----------UCACUGUUUUAAAUUGAA------UACGGGCAUAAAAUGCUUGAUAGGCAUUAUAUGCCUCAUUAGUUUUGCCUGCAUUAAAUAAAAAACACUAGUUAAUUA -----------....((((((..(((.((------(.((((((......(((.(((((((((...)))))).))))))..)))))).))).)))..))))))........... ( -21.00) >DroEre_CAF1 225674 105 - 1 -GCUGGAGAUUUCAGUGGUUUAAAUUGAG------UACGGGCAUAAAACGCCUGAUAGGCAUUAUAUGCCUCAUUAGUUUUCCCUGCAUUAAAUAA-AAGCACUAGUUAAUUA -(((....(((((((.((...(((((((.------...(((((((.((.(((.....))).)).)))))))..))))))).)))))....))))..-.)))............ ( -26.10) >DroYak_CAF1 235645 111 - 1 -GGUGGAGAUUUCAGUGGUUUAAAUUGAAUACGAAUACGGGCAUAAAAUGCUUGAUAGGCAUUAUAUGCCUCAUUAGUUUUCACUGCAUUAAAUAA-AAACACUAGUUAAUUA -(((((((((((((((.......)))))).........(((((((.((((((.....)))))).))))))).....)))))))))...........-................ ( -27.30) >DroAna_CAF1 213410 105 - 1 CGAUAGAGAUAGGCUUGAUUUAAAUCGAU------UAUGAGCAUAAAGCGUUCCAUGGGCAUUGUAUGCCUCAUUAGUUCUUUGUGCAUUAAAAAA-CAGCA-UAGUUAAUUA .(((..((((.......))))..)))(((------((((.((((((((..((..((((((((...))))).))).))..)))))))).........-...))-)))))..... ( -19.72) >consensus __________UUCAGUGGUUUAAAUUGAA______UACGGGCAUAAAAUGCUUGAUAGGCAUUAUAUGCCUCAUUAGUUUUGCCUGCAUUAAAUAA_AAACACUAGUUAAUUA ......................................(((((((.((((((.....)))))).))))))).......................................... (-14.13 = -14.22 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:15 2006