| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,387,942 – 12,388,058 |
| Length | 116 |
| Max. P | 0.934192 |

| Location | 12,387,942 – 12,388,058 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.34 |
| Mean single sequence MFE | -32.98 |
| Consensus MFE | -18.91 |
| Energy contribution | -19.98 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.934192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12387942 116 + 22407834 GUUUUGAUAAGUCUUAUGGCAGCGGCAAGUCGGCUUACAGCAUCAGAAUA-AUGACGCGCACUUAAGCCGCCUUCGCAUGUCCACCC---GACUAUCAAACCACCCACUUUACAAACCAC (.(((((((.(((...(((((((((..((.(((((((..((.(((.....-.))).)).....))))))).)))))).)).)))...---)))))))))).).................. ( -32.70) >DroPse_CAF1 294565 105 + 1 -----GAUAAGUUUUAUGGCAACUGAGAUGCGGCUUACAGCAUCAGCAUAAAUGACGCGCACUUAAGCCGUGUCC-------CGGCC---GACCACCUUGAGGCCCGGACAGGCUCCUAC -----.....((((((.(....)))))))((((((((..((.(((.......))).)).....))))))))((((-------.((((---...........)))).)))).......... ( -33.50) >DroAna_CAF1 199175 110 + 1 GUUUUGAUAAGUUUUAUGGCAACUGCGAGGCGGCUUACAGCAUCAGAAUA-AUGACGCGCACUUAAGCUGCUGCCGCAUCGCCAACCACCAGCCGCCAAACCACC---------AACCAC ................((((...((((.(((((((((..((.(((.....-.))).)).....)))))))))..))))..)))).....................---------...... ( -28.80) >DroPer_CAF1 297001 105 + 1 -----GAUAAGUUUUAUGGGAACUGAGAUGCGGCUUACAGCAUCAGCAUAAAUGACGCGCACUUAAGCCGUGUCC-------CGGCC---GACCACCUUGAGGCCCGGACAGGCUCCUAC -----...........(((((.(((.(((((((((((..((.(((.......))).)).....)))))))))))(-------(((((---...........)).)))).)))..))))). ( -36.90) >consensus _____GAUAAGUUUUAUGGCAACUGAGAGGCGGCUUACAGCAUCAGAAUA_AUGACGCGCACUUAAGCCGCGUCC_______CAGCC___GACCACCAAAACACCCGGACAGGCAACCAC ..........(((((((((.......(((((((((((..((.(((.......))).)).....))))))))))).........((.......)).)))))))))................ (-18.91 = -19.98 + 1.06)

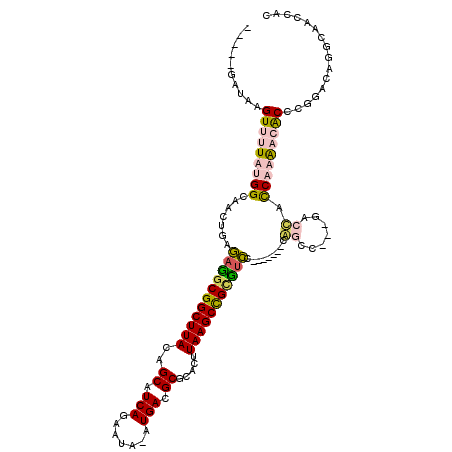
| Location | 12,387,942 – 12,388,058 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.34 |
| Mean single sequence MFE | -40.67 |
| Consensus MFE | -20.17 |
| Energy contribution | -21.18 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12387942 116 - 22407834 GUGGUUUGUAAAGUGGGUGGUUUGAUAGUC---GGGUGGACAUGCGAAGGCGGCUUAAGUGCGCGUCAU-UAUUCUGAUGCUGUAAGCCGACUUGCCGCUGCCAUAAGACUUAUCAAAAC ....................((((((((((---..((((.((.(((.((((((((((.....((((((.-.....))))))..))))))).)))..)))))))))..))).))))))).. ( -41.30) >DroPse_CAF1 294565 105 - 1 GUAGGAGCCUGUCCGGGCCUCAAGGUGGUC---GGCCG-------GGACACGGCUUAAGUGCGCGUCAUUUAUGCUGAUGCUGUAAGCCGCAUCUCAGUUGCCAUAAAACUUAUC----- ...(..((((....))))..)...((((.(---(((.(-------(((..(((((((.....((((((.......))))))..)))))))..)))).))))))))..........----- ( -38.40) >DroAna_CAF1 199175 110 - 1 GUGGUU---------GGUGGUUUGGCGGCUGGUGGUUGGCGAUGCGGCAGCAGCUUAAGUGCGCGUCAU-UAUUCUGAUGCUGUAAGCCGCCUCGCAGUUGCCAUAAAACUUAUCAAAAC .((((.---------(((....((((((((((.(((.(((..((((((.((.((......))))((((.-.....)))))))))).)))))).).)))))))))....))).)))).... ( -44.40) >DroPer_CAF1 297001 105 - 1 GUAGGAGCCUGUCCGGGCCUCAAGGUGGUC---GGCCG-------GGACACGGCUUAAGUGCGCGUCAUUUAUGCUGAUGCUGUAAGCCGCAUCUCAGUUCCCAUAAAACUUAUC----- ...(((((.(((((.((((...........---)))).-------)))))(((((((.....((((((.......))))))..))))))).......))))).............----- ( -38.60) >consensus GUAGGAGCCUGUCCGGGCCGCAAGGUGGUC___GGCCG_______GGACACGGCUUAAGUGCGCGUCAU_UAUGCUGAUGCUGUAAGCCGCAUCGCAGUUGCCAUAAAACUUAUC_____ ...............((((((....((((.....)))).....((((...(((((((.....((((((.......))))))..)))))))..)))).))))))................. (-20.17 = -21.18 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:08 2006