
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,373,977 – 12,374,107 |
| Length | 130 |
| Max. P | 0.942375 |

| Location | 12,373,977 – 12,374,067 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 85.80 |
| Mean single sequence MFE | -22.07 |
| Consensus MFE | -17.24 |
| Energy contribution | -18.30 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12373977 90 + 22407834 AAGCCAUCCCA--ACCACCCACUCCAGCCGUCCUCCAUGUCCACAAACAUGGCCACAAAUGGCCAUCAGUUGUAUCCGGCGAUAACGGCAAC ...........--.............(((((..(((......((((..(((((((....)))))))...)))).....).))..)))))... ( -22.80) >DroVir_CAF1 245995 88 + 1 -CGCCACCCCCGCAGCACCAACUUCAGCAGCA---GCAGCCCACAAACAUGGCCACAAAUGGCCAUCAGUCGUAUCCGGCGAUAACGGCAAC -.(((......((.((..........)).)).---.............(((((((....)))))))..(((((.....)))))...)))... ( -22.30) >DroSec_CAF1 198036 90 + 1 AAGCCAUCCCA--ACCAUCCACUCCAGCCGUCCUCCAUGUCCACAAACAUGGCCACAAAUGGCCAUCAGUUGUAUCCGGCGAUAACGGCAAC ...........--.............(((((..(((......((((..(((((((....)))))))...)))).....).))..)))))... ( -22.80) >DroSim_CAF1 203286 90 + 1 AAGCCAUCCCA--ACCAUCCACUCCAGCCGUCCUCCAUGUCCACAAACAUGGCCACAAAUGGCCAUCAGUUGUAUCCGGCGAUAACGGCAAC ...........--.............(((((..(((......((((..(((((((....)))))))...)))).....).))..)))))... ( -22.80) >DroWil_CAF1 254962 84 + 1 --GUCAUCGCC--CCCUCCGAC----GCCCUCAUCGAUGUCCGCAAACAUGACCACAAAUGGCCAUCAGUUGUAUCCGGCGAUAACAGCGAC --((.((((((--......(((----(.(......).)))).((((..(((.(((....))).)))...))))....)))))).))...... ( -18.90) >DroYak_CAF1 206114 90 + 1 AAGCCAUCCCA--ACCACCCACUCCAGCCGUCCUCCAUGUCCACAAACAUGGCCACAAAUGGCCAUCAGUUGUAUCCGGCGAUAACGGCAAC ...........--.............(((((..(((......((((..(((((((....)))))))...)))).....).))..)))))... ( -22.80) >consensus AAGCCAUCCCA__ACCACCCACUCCAGCCGUCCUCCAUGUCCACAAACAUGGCCACAAAUGGCCAUCAGUUGUAUCCGGCGAUAACGGCAAC ..........................(((((..((...(((.((((..(((((((....)))))))...))))....)))))..)))))... (-17.24 = -18.30 + 1.06)

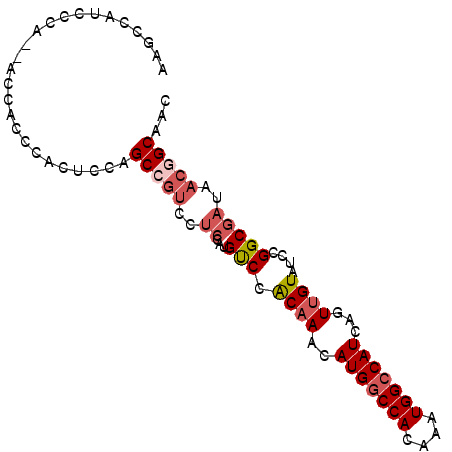
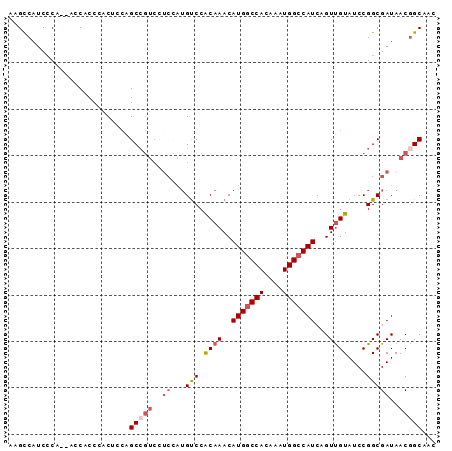
| Location | 12,373,990 – 12,374,107 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.35 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -21.69 |
| Energy contribution | -22.25 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.767466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12373990 117 + 22407834 CACCCACUCCAGCCGUCCUCCAUGUCCACAAACAUGGCCACAAAUGGCCAUCAGUUGUAUCCGGCGAUAACGGCAACGUAUUGGCUCCCGCCGCCGAAUCCGGCGCCAUAUUUAGUU ...........((((............((((..(((((((....)))))))...))))...((((....(((....)))...(((....)))))))....))))............. ( -36.70) >DroVir_CAF1 246009 114 + 1 CACCAACUUCAGCAGCA---GCAGCCCACAAACAUGGCCACAAAUGGCCAUCAGUCGUAUCCGGCGAUAACGGCAACGUAUUGGCUGCCGCCACCAAAUCCGGCGCCAUAUUUAGUU ....((((......((.---((((((.......(((((((....)))))))..(((((.....))))).(((....)))...)))))).(((.........))))).......)))) ( -35.72) >DroGri_CAF1 252910 97 + 1 --------------------ACAGUCCACAAACAUGGCCACAAAUGGCCAUCAGUCGUAUCCGGCGAUAACGGCAACGUAUUGGCUACCGCCACCAAAUCCGGCGCCAUAUUUAGUU --------------------.........(((.(((((......((((((...(((((.....))))).(((....)))..))))))..(((.........)))))))).))).... ( -27.30) >DroWil_CAF1 254973 113 + 1 CUCCGAC----GCCCUCAUCGAUGUCCGCAAACAUGACCACAAAUGGCCAUCAGUUGUAUCCGGCGAUAACAGCGACCUAUUGGCUACCCCCACCGAAUCCGGCGCCAUAUUUAGUU ....(.(----(((....(((((((......)))).........((((((...((((((((....))).))))).......)))))).......)))....)))).).......... ( -22.60) >DroMoj_CAF1 250797 97 + 1 --------------------GCAGCCCACUAACAUGGCCACAAAUGGCCAUCAGUCGUAUCCGGCGAUAACGGCAACGUAUUGGCUGCCGCCACCAAAUCCGGCGCCAUAUUUAGUU --------------------((((((.......(((((((....)))))))..(((((.....))))).(((....)))...))))))((((.........))))............ ( -34.10) >DroAna_CAF1 185452 117 + 1 CACCAGCACCACACCUCGCCAGUGUCCACAAGCAUGACCACAAAUGGCCAUCAGUCGUAUCCGGCGAUAACGGCAACGUAUUGGCUGCCGCCACCGAAUCCGGCGCCAUAUUUAGUU ...............(((((.(((..((......))..)))..(((((.....)))))....)))))..(((....)))..((((.((((..........))))))))......... ( -30.20) >consensus CACC_AC____GCC_UC___GCAGUCCACAAACAUGGCCACAAAUGGCCAUCAGUCGUAUCCGGCGAUAACGGCAACGUAUUGGCUGCCGCCACCAAAUCCGGCGCCAUAUUUAGUU ..............................((((((((((....)))))))...........((((...(((....)))..((((....))))((......)))))).......))) (-21.69 = -22.25 + 0.56)

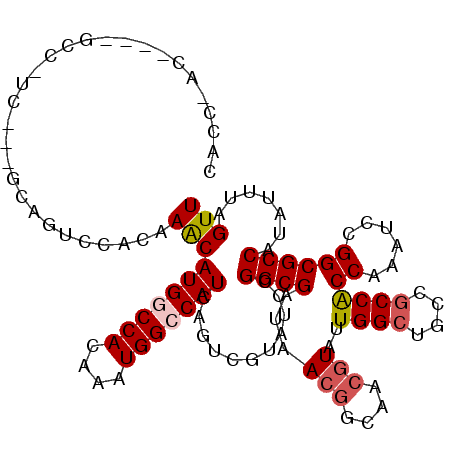
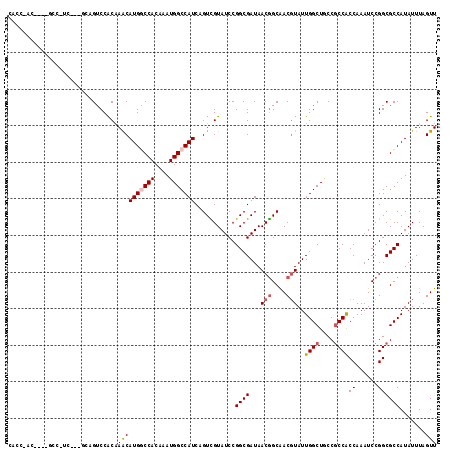
| Location | 12,373,990 – 12,374,107 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.35 |
| Mean single sequence MFE | -44.05 |
| Consensus MFE | -36.20 |
| Energy contribution | -36.20 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12373990 117 - 22407834 AACUAAAUAUGGCGCCGGAUUCGGCGGCGGGAGCCAAUACGUUGCCGUUAUCGCCGGAUACAACUGAUGGCCAUUUGUGGCCAUGUUUGUGGACAUGGAGGACGGCUGGAGUGGGUG .(((.........((((....))))(((....)))..(((...((((((....(((..(((((...(((((((....)))))))..)))))....)))..))))))....)))))). ( -43.20) >DroVir_CAF1 246009 114 - 1 AACUAAAUAUGGCGCCGGAUUUGGUGGCGGCAGCCAAUACGUUGCCGUUAUCGCCGGAUACGACUGAUGGCCAUUUGUGGCCAUGUUUGUGGGCUGC---UGCUGCUGAAGUUGGUG ............((((((.((..(..(((((((((....(((..(((.......)))..)))....(((((((....))))))).......))))))---)))..)..)).)))))) ( -50.90) >DroGri_CAF1 252910 97 - 1 AACUAAAUAUGGCGCCGGAUUUGGUGGCGGUAGCCAAUACGUUGCCGUUAUCGCCGGAUACGACUGAUGGCCAUUUGUGGCCAUGUUUGUGGACUGU-------------------- ...((((((((((.((((....((((((((((((......)))))))))))).)))).(((((.((.....)).)))))))))))))))........-------------------- ( -38.00) >DroWil_CAF1 254973 113 - 1 AACUAAAUAUGGCGCCGGAUUCGGUGGGGGUAGCCAAUAGGUCGCUGUUAUCGCCGGAUACAACUGAUGGCCAUUUGUGGUCAUGUUUGCGGACAUCGAUGAGGGC----GUCGGAG .........((((((((.(((((((((.((..(((....)))..))....))))))))).)..((.(((((((....)))))(((((....)))))..)).)))))----))))... ( -38.90) >DroMoj_CAF1 250797 97 - 1 AACUAAAUAUGGCGCCGGAUUUGGUGGCGGCAGCCAAUACGUUGCCGUUAUCGCCGGAUACGACUGAUGGCCAUUUGUGGCCAUGUUAGUGGGCUGC-------------------- ..........(((.((((....((((((((((((......)))))))))))).))))...(.(((((((((((....))))))..))))).))))..-------------------- ( -42.60) >DroAna_CAF1 185452 117 - 1 AACUAAAUAUGGCGCCGGAUUCGGUGGCGGCAGCCAAUACGUUGCCGUUAUCGCCGGAUACGACUGAUGGCCAUUUGUGGUCAUGCUUGUGGACACUGGCGAGGUGUGGUGCUGGUG ..........(((((((.((((((((((((((((......))))))))))))(((((.(((((...(((((((....)))))))..)))))....))))))).)).))))))).... ( -50.70) >consensus AACUAAAUAUGGCGCCGGAUUCGGUGGCGGCAGCCAAUACGUUGCCGUUAUCGCCGGAUACGACUGAUGGCCAUUUGUGGCCAUGUUUGUGGACAGC___GA_GGC____GU_GGUG ...((((((((((.((((...(((((((((((((......))))))))))))))))).(((((.((.....)).)))))))))))))))............................ (-36.20 = -36.20 + 0.00)

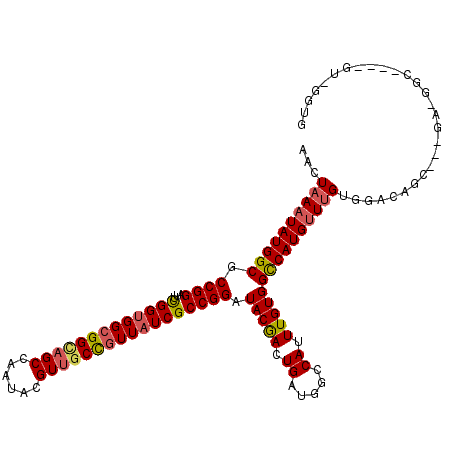
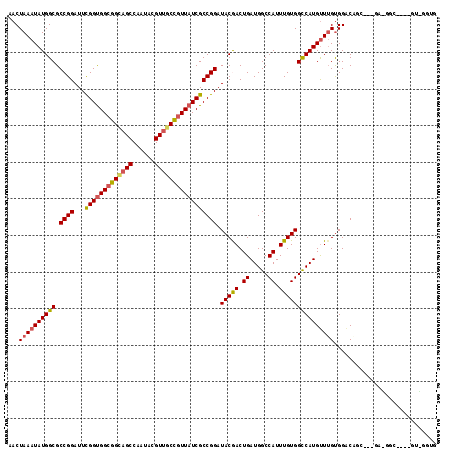
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:03 2006