
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,372,018 – 12,372,123 |
| Length | 105 |
| Max. P | 0.658398 |

| Location | 12,372,018 – 12,372,123 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 83.46 |
| Mean single sequence MFE | -20.08 |
| Consensus MFE | -14.22 |
| Energy contribution | -14.42 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12372018 105 - 22407834 C--CCCCGCCCUUUCCCACCCCUUGCCACUUGUCUCUACUGUCUAUUUUCAAUUUCUUCUGACCCAGCUGAAAUUGACGUUUGCCAGUCUAUUGACAGACUGCCUAA----- .--.....................((...(((((...((((.(.....((((((((..(((...)))..)))))))).....).)))).....)))))...))....----- ( -17.90) >DroVir_CAF1 243541 103 - 1 CAUUUCUAUGCU-UUCCACAACUUGUCGG-------GUCUGUCUAUUUUCAAUUUCUUGUGGCCCGGCU-AAAUUGACGUUUGCCAGUCUAUUGACAGACUGCCUCUUCUUC .........((.-.....(((.(((((((-------(((.....................)))))))).-)).)))......))((((((......)))))).......... ( -20.90) >DroSec_CAF1 196101 103 - 1 C--CUCCCGGCU-UCCC-CCCUUUGCCACCUGAUUCGACUGUCUAUUUUCAAUUUCUUCUGACCCAGCUGAAAUUGACGUUUGCCAGUCUAUUGACAGACUGCCUAA----- .--.....(((.-....-......)))..(((....(((((.(.....((((((((..(((...)))..)))))))).....).)))))......))).........----- ( -21.00) >DroSim_CAF1 201350 104 - 1 C--CUCCCGGCUUUCCC-CCCUUUGCCACCUGAUUCGACUGUCUAUUUUCAAUUUCUUCUGACCCAGCUGAAAUUGACGUUUGCCAGUCUAUUGACAGACUGCCUAA----- .--.....(((......-......)))..(((....(((((.(.....((((((((..(((...)))..)))))))).....).)))))......))).........----- ( -20.90) >DroEre_CAF1 194875 105 - 1 C--CUCCGAACUUUUCCCCCGCUUUCCACCUGACUCGACUGUCUAUUUUCAAUUUCUUCUGACCCAGCUGAAAUUGACGUUUGCCAGUCUAUUGACAGACUGCCUAA----- .--...(((((....................(((......))).....((((((((..(((...)))..)))))))).))))).((((((......)))))).....----- ( -21.40) >DroYak_CAF1 204213 102 - 1 C--AUCCCGACUUUUCCCCC---UUCCACCUGACUCGACUGUCUAUUUUCAAUUUCUUCUGACCCAGCUGAAAUUGACGUUUGCCAGUCUAUUGACAGACUGCCUAG----- .--....((...........---........(((......))).....((((((((..(((...)))..))))))))))...(.((((((......)))))).)...----- ( -18.40) >consensus C__CUCCCGGCUUUCCC_CCCCUUGCCACCUGACUCGACUGUCUAUUUUCAAUUUCUUCUGACCCAGCUGAAAUUGACGUUUGCCAGUCUAUUGACAGACUGCCUAA_____ ........................................((((....((((((((..(((...)))..)))))))).........(((....)))))))............ (-14.22 = -14.42 + 0.20)

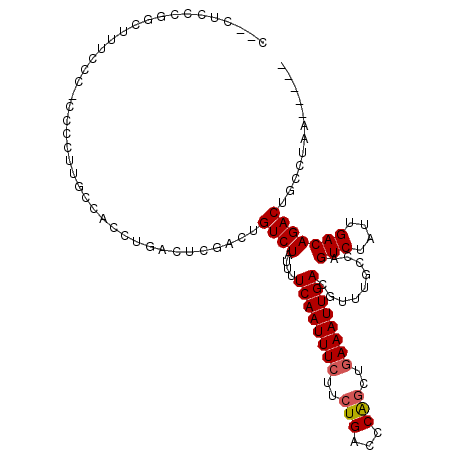
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:15:00 2006