
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,368,943 – 12,369,044 |
| Length | 101 |
| Max. P | 0.649061 |

| Location | 12,368,943 – 12,369,044 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 81.74 |
| Mean single sequence MFE | -21.08 |
| Consensus MFE | -14.44 |
| Energy contribution | -14.95 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
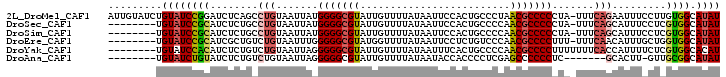
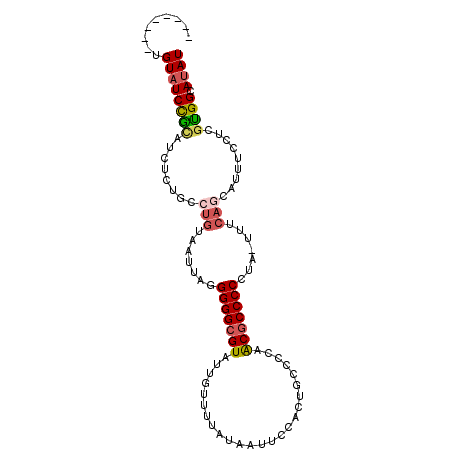
>2L_DroMel_CAF1 12368943 101 + 22407834 AUUGUAUCUGUAUCCGGAUCUCAGCCUGUAAUUAUGGGGCGUAUUGUUUUAUAAUUCCACUGCCCUAACGCCCCCUA-UUUCAGAAUUUCCUUGUGGCAUAU ...(((((..((...(((.......(((.(((...(((((((...((..............))....)))))))..)-)).)))....))).))..).)))) ( -18.84) >DroSec_CAF1 192994 93 + 1 --------UGUAUCCGCAUCUCUGCCUGUAAUUAUGGGGCGUAUUGUUUUAUAAUUCCACUGCCCCAACGCCCCCUA-UUUCAGCAUUUCCUCGUGGCAUAU --------.((((((((.....(((.((.(((...(((((((...((..............))....)))))))..)-)).))))).......)))).)))) ( -20.34) >DroSim_CAF1 198240 93 + 1 --------UGUAUCCGCAUCUCUGCCUGUAAUUAUGGGGCGUAUUGUUUUAUAAUUCCACUGCCCCAACGCCCCCUA-UUUCAGCAUUUCCUCGUGGCAUAU --------.((((((((.....(((.((.(((...(((((((...((..............))....)))))))..)-)).))))).......)))).)))) ( -20.34) >DroEre_CAF1 191722 93 + 1 --------UGUAUCCGCAUCGCUGUCUGUAAUUUGGGGGCGUAUGGUUUUAUAAUUCCUCUGUCCCAACGCCCCUUU-UUUCAACAUUUGCUGGUGGCAUAU --------.((((((((...(((((.((......((((((((.(((..(............)..)))))))))))..-...)))))...))..)))).)))) ( -25.70) >DroYak_CAF1 201052 94 + 1 --------UGUAUCCACAUCUCUGUCUGUAAUUAGGGGGCGUAUUGUUUUAUAAUUUCACUGCCCCAACGCCCCUUUUUUUCACCAUUUUCUCGUGGCACAU --------(((..((((.........((.((..(((((((((...((..............))....)))))))))..)).))..........)))).))). ( -19.35) >DroAna_CAF1 180531 86 + 1 --------UGUAUCUGUAUCUCUGUCUGUAAUUAGGGGGCGUAUUGUUUUAUAAUACCACCCCUCGAGCCCCCCUC-------GCACUU-GUUGCGGCAUAU --------..............((.(((((((.((((((.(((((((...)))))))..)))).((((.....)))-------)..)).-)))))))))... ( -21.90) >consensus ________UGUAUCCGCAUCUCUGCCUGUAAUUAGGGGGCGUAUUGUUUUAUAAUUCCACUGCCCCAACGCCCCCUA_UUUCAGCAUUUCCUCGUGGCAUAU .........((((((((........(((.......(((((((.........................))))))).......))).........)))).)))) (-14.44 = -14.95 + 0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:59 2006