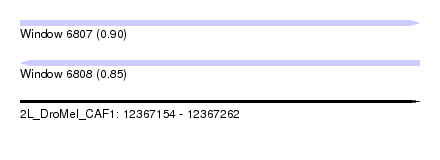
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,367,154 – 12,367,262 |
| Length | 108 |
| Max. P | 0.896151 |

| Location | 12,367,154 – 12,367,262 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -14.60 |
| Energy contribution | -15.30 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
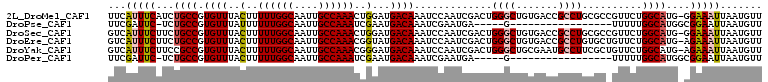
>2L_DroMel_CAF1 12367154 108 + 22407834 AACAUUAAUUUCC-CAUGCCAGAACGGCGCAGGCGGUCACAGCCCAGUCGAUUGGAUUUGUCAUCCAGUUUGGCAAUUGCCAAAAAGUAAACACGGCAGAUGAAAUGAA .......(((((.-..((((....((((...(((.......)))..))))(((((((.....)))))))(((((....)))))...........))))...)))))... ( -31.60) >DroPse_CAF1 268321 86 + 1 AACAUUAAUUCCGCCAUGCCAAAAA-----------------C-----UCAUUCGAUUUGUCAUUCGAUUUGGCAAUUGCCAAAAAAUAAACACGGCAGA-GAAUCGAA ....(..((((.....((((.....-----------------.-----....((((........))))((((((....))))))..........))))..-))))..). ( -18.80) >DroSec_CAF1 191195 108 + 1 AACAUUAAUUUCC-CAUGCCAGAACGGCGCAGGCGGUCACAGCCCAGUCGAUUGGAUUUGUCAUCCAGUUUGGCAAUUGCCAAAAAGUAAACACGGCAGAAGAAAUGAC .......(((((.-..((((....((((...(((.......)))..))))(((((((.....)))))))(((((....)))))...........))))...)))))... ( -31.60) >DroEre_CAF1 189877 108 + 1 AACAUUAAUUUCU-CAUGCCAGAACAGCACAGGCGGUCACAGCCCAGUCGAUUGGAUUUGUCAUACCGUUUGGCAAUUGCCAAAAAGUAAACACGGCAGAAGAAAUGAC .......((((((-..((((......((.((((((((.((((.((((....))))..))))...))))))))))..((((......))))....))))..))))))... ( -30.50) >DroYak_CAF1 199278 108 + 1 AACAUUAAUUUCU-CAUGCCAGAACAGCGAAGGCAUUCGCAGCCCAGUCGAUUGGAUUUGUCAUCCCGUUUGGCAAUUGCCAAAAAGUAAACACGGCGGAAGAAAUGAC .......((((((-..((((......(((((....)))))......((..(((((((.....))))..((((((....)))))).)))..))..))))..))))))... ( -28.90) >DroPer_CAF1 272580 86 + 1 AACAUUAAUUCCGCCAUGCCAAAAA-----------------C-----UCAUUCGAUUUGUCAUUCGAUUUGGCAAUUGCCAAAAAGUAAACACGGCAGA-GAAUCGAA ....(..((((.....((((.....-----------------.-----....((((........))))((((((....))))))..........))))..-))))..). ( -18.80) >consensus AACAUUAAUUUCC_CAUGCCAGAAC_GCGCAGGCGGUCACAGCCCAGUCGAUUGGAUUUGUCAUCCAGUUUGGCAAUUGCCAAAAAGUAAACACGGCAGAAGAAAUGAA ................((((...........(((.......))).......((((..((((((.......))))))...))))...........))))........... (-14.60 = -15.30 + 0.70)


| Location | 12,367,154 – 12,367,262 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -29.27 |
| Consensus MFE | -17.45 |
| Energy contribution | -17.95 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
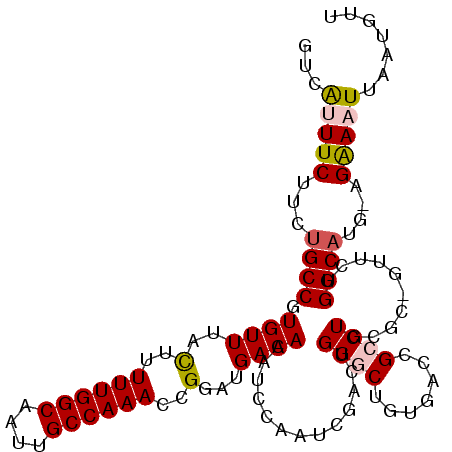
>2L_DroMel_CAF1 12367154 108 - 22407834 UUCAUUUCAUCUGCCGUGUUUACUUUUUGGCAAUUGCCAAACUGGAUGACAAAUCCAAUCGACUGGGCUGUGACCGCCUGCGCCGUUCUGGCAUG-GGAAAUUAAUGUU ......((((..(((..(((.....((((((....)))))).(((((.....)))))...)))..))).))))...(((..(((.....)))..)-))........... ( -28.50) >DroPse_CAF1 268321 86 - 1 UUCGAUUC-UCUGCCGUGUUUAUUUUUUGGCAAUUGCCAAAUCGAAUGACAAAUCGAAUGA-----G-----------------UUUUUGGCAUGGCGGAAUUAAUGUU ........-(((((((((((.((((((((((....))))))((((........))))..))-----)-----------------)....)))))))))))......... ( -25.70) >DroSec_CAF1 191195 108 - 1 GUCAUUUCUUCUGCCGUGUUUACUUUUUGGCAAUUGCCAAACUGGAUGACAAAUCCAAUCGACUGGGCUGUGACCGCCUGCGCCGUUCUGGCAUG-GGAAAUUAAUGUU ...((((((..((((..........((((((....)))))).(((((.....)))))...((.(((((.((....))..)).))).)).))))..-))))))....... ( -30.80) >DroEre_CAF1 189877 108 - 1 GUCAUUUCUUCUGCCGUGUUUACUUUUUGGCAAUUGCCAAACGGUAUGACAAAUCCAAUCGACUGGGCUGUGACCGCCUGUGCUGUUCUGGCAUG-AGAAAUUAAUGUU ...(((((((.((((.((((((((.((((((....)))))).)))).)))).............((((.......))))..........)))).)-))))))....... ( -31.10) >DroYak_CAF1 199278 108 - 1 GUCAUUUCUUCCGCCGUGUUUACUUUUUGGCAAUUGCCAAACGGGAUGACAAAUCCAAUCGACUGGGCUGCGAAUGCCUUCGCUGUUCUGGCAUG-AGAAAUUAAUGUU ...(((((((..(((.((((..((.((((((....)))))).))...)))).............((((.(((((....))))).)))).)))..)-))))))....... ( -31.50) >DroPer_CAF1 272580 86 - 1 UUCGAUUC-UCUGCCGUGUUUACUUUUUGGCAAUUGCCAAAUCGAAUGACAAAUCGAAUGA-----G-----------------UUUUUGGCAUGGCGGAAUUAAUGUU ........-(((((((((((.((((((((((....))))))((((........))))..))-----)-----------------)....)))))))))))......... ( -28.00) >consensus GUCAUUUCUUCUGCCGUGUUUACUUUUUGGCAAUUGCCAAACCGGAUGACAAAUCCAAUCGACUGGGCUGUGACCGCCUGCGC_GUUCUGGCAUG_AGAAAUUAAUGUU ...(((((...((((.((((..(..((((((....))))))..)...)))).............((((.......))))..........))))....)))))....... (-17.45 = -17.95 + 0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:58 2006