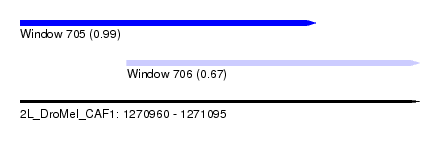
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,270,960 – 1,271,095 |
| Length | 135 |
| Max. P | 0.985415 |

| Location | 1,270,960 – 1,271,060 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 87.13 |
| Mean single sequence MFE | -25.58 |
| Consensus MFE | -21.59 |
| Energy contribution | -21.37 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.49 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1270960 100 + 22407834 CCUCCACCAAAUGCACUACACACAGCACUAUUUGAUGCAUUUUGCAUAAGCGCUGGGCAAAUGCCUUUGCUGCUUUUAUGCAAAUUUCUAGCAUUUUCCA ........((((((..........((((.....).)))..(((((((((((((.((((....))))..)).)))..))))))))......)))))).... ( -26.60) >DroPse_CAF1 44366 100 + 1 CCGCCUUCUUCUGUGGCACCCCUUCCACUAUUUGAUGCAUUUUGCAUAAGCGUUGGGCAAAUGCCUUUGCUGCUUUUAUGCAAAUUUCUAGCAUUUUCCA ..(((.........)))................(((((..(((((((((((((.((((....))))..)).)))..))))))))......)))))..... ( -24.60) >DroSec_CAF1 30539 99 + 1 CCU-CACCAAAUGCACUACACACAGCACUAUUUGAUGCAUUUUGCAUAAGCGCUGGGCAAAUGCCUUUGCUGCUUUUAUGCAAAUUUCUAGCAUUUUCCA ...-....((((((..........((((.....).)))..(((((((((((((.((((....))))..)).)))..))))))))......)))))).... ( -26.60) >DroEre_CAF1 43115 100 + 1 CCUCCACCAAAUGCACUGCACACAGCACUAUUUGAUGCAUUUUGCAUAAGCGCUGGGCAAAUGCCUUUGCUGCUUUUAUGCAAAUUUCUAGCAUUUUCCA ........((((((..((((..(((......))).)))).(((((((((((((.((((....))))..)).)))..))))))))......)))))).... ( -26.80) >DroAna_CAF1 54794 90 + 1 CGUCCAC----------GCCCCCUACACUAUUUGAUGCAUUUUGCAUAAGCGCUGGGCAAAUGCCUUUGCUGCUUUUAUGCAAAUUUCUUGCAUUUUCCA .......----------................((((((.(((((((((((((.((((....))))..)).)))..)))))))).....))))))..... ( -24.30) >DroPer_CAF1 44942 100 + 1 CCGCCUUCUUCUGUGGCACCCCUUCCACUAUUUGAUGCAUUUUGCAUAAGCGUUGGGCAAAUGCCUUUGCUGCUUUUAUGCAAAUUUCUAGCAUUUUCCA ..(((.........)))................(((((..(((((((((((((.((((....))))..)).)))..))))))))......)))))..... ( -24.60) >consensus CCUCCACCAAAUGCACUACACACAGCACUAUUUGAUGCAUUUUGCAUAAGCGCUGGGCAAAUGCCUUUGCUGCUUUUAUGCAAAUUUCUAGCAUUUUCCA .................................(((((..(((((((((((((.((((....))))..)).)))..))))))))......)))))..... (-21.59 = -21.37 + -0.22)


| Location | 1,270,996 – 1,271,095 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.48 |
| Mean single sequence MFE | -22.13 |
| Consensus MFE | -12.15 |
| Energy contribution | -12.49 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1270996 99 + 22407834 GCAUUUUGCAUAAGCGCU--GGGCAAAUGCCUUUGC---UGCUU-------UUAUGCAAAUU-UCUAGCAUUUUC---CAUGUCGUUACAUUCUUC---GUCGUGUUUUAG-CUGCGA-C ((..(((((((((((((.--((((....))))..))---.))).-------.))))))))..-...(((......---((((.((..........)---).)))).....)-))))..-. ( -26.80) >DroPse_CAF1 44402 103 + 1 GCAUUUUGCAUAAGCGUU--GGGCAAAUGCCUUUGC---UGCUU-------UUAUGCAAAUU-UCUAGCAUUUUC---CAUGUCGUUACAUUCUUGUUCCUUGUGUUUUCCAUUCUGC-C (((.(((((((((((((.--((((....))))..))---.))).-------.))))))))..-............---((((..(..(((....)))..).))))..........)))-. ( -20.50) >DroGri_CAF1 48928 101 + 1 GCAUUUUGCAUAAGCGCU--UGG-AAAUGCCUUUGU---CGUUUUAGCAAUUUAUGCAAAUU-UCGUGCAUUUUACCCCAUGUUGUUACAUUCUUU---GU-AUGUCUUAG--------C ((((((((((((((.(((--..(-((((((....).---)))))))))..))))))))))..-..))))............((((..((((.....---..-))))..)))--------) ( -19.60) >DroWil_CAF1 43311 109 + 1 GCAUUUUGCAUAAGCGCUUUUGGAAAAUGCCUUUGCCUUUGCUU-------UUAUGCAAAUUUUCGUGCAUUUUC---CAUGUCGUUACAUUCUUGCCUUUUAUGUUUAUACUUAUAU-A ((((...(((..(((((...((((((((((.......(((((..-------....))))).......))))))))---)).).)))).......))).....))))............-. ( -22.44) >DroAna_CAF1 54820 100 + 1 GCAUUUUGCAUAAGCGCU--GGGCAAAUGCCUUUGC---UGCUU-------UUAUGCAAAUU-UCUUGCAUUUUC---CAUGUCGUUACAUUCUUG---GUCGUGUUUUAG-UUUUGCCC (((.(((((((((((((.--((((....))))..))---.))).-------.))))))))..-.((.((((...(---((..............))---)..))))...))-...))).. ( -24.24) >DroPer_CAF1 44978 103 + 1 GCAUUUUGCAUAAGCGUU--GGGCAAAUGCCUUUGC---UGCUU-------UUAUGCAAAUU-UCUAGCAUUUUC---CAUGUCGUUACAUUCUUCUUCCUUGUGUUUUCCAUUCUGC-C (((.(((((((((((((.--((((....))))..))---.))).-------.))))))))..-....((((....---.))))................................)))-. ( -19.20) >consensus GCAUUUUGCAUAAGCGCU__GGGCAAAUGCCUUUGC___UGCUU_______UUAUGCAAAUU_UCUAGCAUUUUC___CAUGUCGUUACAUUCUUG___GUUGUGUUUUAG_UUCUGC_C (((((((((((((.......(((((...((....))...))))).......))))))))).......((((........))))...................)))).............. (-12.15 = -12.49 + 0.34)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:09 2006