
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,355,144 – 12,355,247 |
| Length | 103 |
| Max. P | 0.562900 |

| Location | 12,355,144 – 12,355,247 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 81.75 |
| Mean single sequence MFE | -14.12 |
| Consensus MFE | -8.16 |
| Energy contribution | -8.61 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12355144 103 - 22407834 AUUGAUGCCAUUUAGGAAAUUACUUCCAAUUUACUUUAAAAUUUCUUCAUUUUCCAAUUUCCUGCGUAAGUAAUACCAGCGCUUGCAGCCGCGCCAUCCAUCU------------- ...((((.......(((((........(((((......))))).......))))).......(((....)))......((((........))))....)))).------------- ( -15.16) >DroPse_CAF1 253315 88 - 1 AAUGAUGCCAUUUAGGAAAUUACUUCCAAUUUACUUUUAAAUUUCUUCAUUUUCCAAUUUCCUGCGUAAGUAAUUUCCCCA---------------UUCGCCC------------- ......((......((((((((((((((..........((((......))))..........)).).)))))))))))...---------------...))..------------- ( -11.35) >DroEre_CAF1 177901 101 - 1 AACGAUGCCAUUUAGGAAAUUACUUCCCAUUUACUUUUAAAUUUCUUCAUUUUCCAAUUUCCUGCGUAAGUAAUUCCAGCGCUUGCAGCC--GCCAUCCAUCU------------- ...((((.(....((((((((..................................))))))))((((((((.........)))))).)).--).)))).....------------- ( -13.88) >DroYak_CAF1 185354 101 - 1 AACGAUGCCAUUUAGGAAAUUACUUCCAAUUUACUUUUAAAUUUCUUCAUUUUCCAAUUUCCUGCGUAAGUAAUUCCAGCGCUUGCAGCC--GCCAUCCAUCU------------- ...((((.(.....(((((........((((((....)))))).......)))))......(((((...((.......))...)))))..--).)))).....------------- ( -14.56) >DroAna_CAF1 166879 112 - 1 AACGAUGCCAUUUAGGAAAUUACUUUCAAUUUACUUUUAAAUUUCUUCAUUUUCCAAUUUCCUGCGAAAGUAAUUUCAGCGCU-GCAGCC---CCAUUCAUCUUCCCCAUCCCAUU ...((((........((((((((((((((((((....))))))...........((......)).)))))))))))).((...-...)).---.....)))).............. ( -16.80) >DroPer_CAF1 261267 88 - 1 AAUGAUGCCAUUUAGGAAAUUACUUCCAAUUUACUUUUAAAUUUCUUCAUUUUCCAAUUUCCUGCGUAAGUAAUUUCCUCA---------------UUCGCCC------------- ......((.....(((((((((((((((..........((((......))))..........)).).))))))))))))..---------------...))..------------- ( -12.95) >consensus AACGAUGCCAUUUAGGAAAUUACUUCCAAUUUACUUUUAAAUUUCUUCAUUUUCCAAUUUCCUGCGUAAGUAAUUCCAGCGCU_GCAGCC___CCAUCCAUCU_____________ ...((((.......(((((........((((((....)))))).......))))).......(((....)))..........................)))).............. ( -8.16 = -8.61 + 0.45)

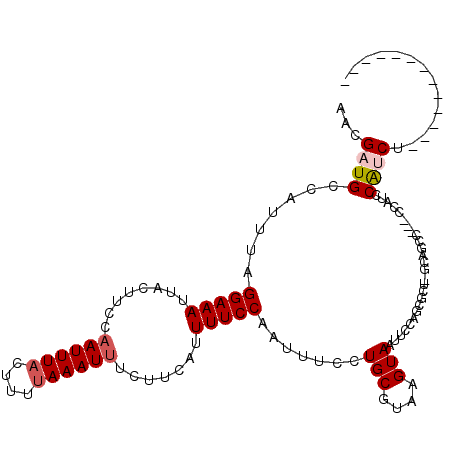
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:56 2006