
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,353,918 – 12,354,017 |
| Length | 99 |
| Max. P | 0.764139 |

| Location | 12,353,918 – 12,354,017 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 79.63 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -19.67 |
| Energy contribution | -18.75 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
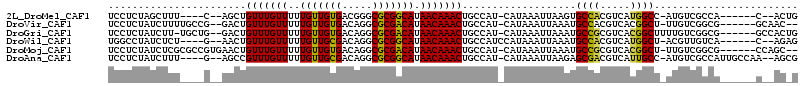
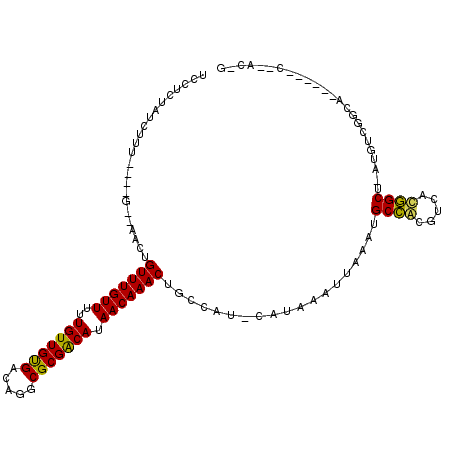
>2L_DroMel_CAF1 12353918 99 + 22407834 UCCUCUAGCUUU----C--AGCUGUUUGUUUUUGUUGUGACGGGCGCGGCAUAACAAACUGCCAU-CAUAAAUUAAGUGCCACGUCAUGGCC-AUGUCGCCA------C--ACUG .....((((...----.--.)))).........((..((((((((((((((........))))..-.(.....)..))))).)))))..)).-.........------.--.... ( -26.60) >DroVir_CAF1 221844 103 + 1 UCCUCUAUCUUUUGCCG--GACUGUUUGUUUUUGUUGUGACAGGCGCGACAUAACAAACUGCCAU-CAUAAAUUAAAUGCCACGUCACGGCU-UUGUCGGCG------GCAAC-- ...........((((((--(...(((((((..(((((((.....))))))).)))))))..))..-............((((((........-.))).))))------)))).-- ( -30.20) >DroGri_CAF1 227233 105 + 1 UCCUCUAUCUU-UGCUG--GACUGUUUGUUUUUGUUGUGACAGGCGCGACAUAACAAACUGCCAU-CAUAAAUUAAAUGCCGCGUCACGGCUUUUGUCGGCG------GCCACUG ...........-((.((--(...(((((((..(((((((.....))))))).)))))))..))).-))..........(((((....((((....)))))))------))..... ( -31.80) >DroWil_CAF1 225355 100 + 1 UGGCCUAUCUCU----G--AACUGUUUGUUUUUGUUGCGACAGGCGCGGCAUAACAAACUGCCAUCCAUAAAUUAAAUGCCACGUCAUGGCU-ACGUUGUCA------C--AGAG ........((((----(--....(((((((..(((((((.....))))))).)))))))...................((((.....)))).-.........------)--)))) ( -26.00) >DroMoj_CAF1 226843 105 + 1 UCCUCUAUCUCGCGCCGUGAACUGUUUGUUUUUGUUGUGACAGGCGCGACAUAACAAACUGCCAU-CAUAAAUUAAAUGCCGCGUCACGGCU-UUGUCGGCG------CCAGC-- ........((.(((((((((...(((((((..(((((((.....))))))).))))))).....)-)))....((((.((((.....)))))-)))..))))------).)).-- ( -33.60) >DroAna_CAF1 163771 105 + 1 UCCUCUAUCUUU----G--AGCCGUUUGUUUUUGUUGCGACAGGCGCGGCAUAACAAACUGCCAU-CAUAAAUUAAGAGCGACGUCAUUGCC-AUGUCGCCAUUGCCAA--AGCG ........((((----(--.((.(((((((..(((((((.....))))))).))))))).))...-..........(.(((((((.......-)))))))).....)))--)).. ( -30.60) >consensus UCCUCUAUCUUU____G__AACUGUUUGUUUUUGUUGUGACAGGCGCGACAUAACAAACUGCCAU_CAUAAAUUAAAUGCCACGUCACGGCU_AUGUCGGCA______C__AC_G .......................(((((((..(((((((.....))))))).)))))))...................((((.....))))........................ (-19.67 = -18.75 + -0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:55 2006