| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,342,079 – 12,342,176 |
| Length | 97 |
| Max. P | 0.583381 |

| Location | 12,342,079 – 12,342,176 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 77.45 |
| Mean single sequence MFE | -25.62 |
| Consensus MFE | -16.12 |
| Energy contribution | -15.99 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12342079 97 + 22407834 ----------GGUUUCUUAAGCCGCCAUUCAUCAGUUUCGA--CUGGCCAAGUAUUUAAGCCAUUUUCUCUAAUGAAUGCGGUCGCGCAUUUUUAAGCGCCC--------ACAAAAC ----------((........(((((.((((((.((....((--.((((...........)))).))...)).))))))))))).((((........))))))--------....... ( -25.40) >DroVir_CAF1 206104 93 + 1 --------ACUAGUUCUUAAGCUGCCACACAACAUUUGCGG-----GCCAAGUAUUUAAGCCAUUUGCGCUAAUGAAUGCGGUCGCGCAUUUUUAAGCGUAC--------CCAA--- --------.......(((((((((((.(.((.....)).))-----))..)))..))))).....((((((...(((((((....)))))))...)))))).--------....--- ( -22.50) >DroPse_CAF1 240375 96 + 1 ------AGACUGGCUCUUAAGCUGCCAUUCAGCAAUUUCGA--CUGGCCAAGUAUUUAAGCCAUUUGCACUAAUGAAUGCGGUCGCGCAUUUUUAAGCGCC-------------AAU ------....((((.(((((((((.....)))).....(((--.((((...........)))).))).......(((((((....)))))))))))).)))-------------).. ( -25.80) >DroWil_CAF1 206692 101 + 1 ------CUUCUACUACUUAAGCUGCCAUUCAGCGAUUUUGGUUCUGGCCAAGUAUUUAAGCCAUUUGCACUAAUGAAUGCGGUCGCGCAUUUUUAAGCGUUC--------AA--AAU ------.........(((((((((.....))))...((((((....))))))...)))))......((.((...(((((((....)))))))...)).))..--------..--... ( -20.20) >DroAna_CAF1 152009 106 + 1 ---------GGGGCUCUUAAGCUGCCAUUCGCCACUUUCGA--CUGGCCAAGUAUUUAAGCCAUUUGCACUAAUGAAUGCGGUCGCGCAUUUUUGAGUGCCCCAUUGCCUACGAAAC ---------((((((.....(((((((.(((.......)))--.))))..))).....))))....(((((...(((((((....)))))))...))))).)).............. ( -32.40) >DroPer_CAF1 246690 102 + 1 GUAAGGGGACUGGCUCUUAAGCUGCCAUUCAGCAAUUUCGA--CUGGCCAAGUAUUUAAGCCAUUUGCACUAAUGAAUGCGGUCGCGCAUUUUUAAGCGCC-------------AAU ...((....))(((.(((((((((.....)))).....(((--.((((...........)))).))).......(((((((....)))))))))))).)))-------------... ( -27.40) >consensus ________ACUGGCUCUUAAGCUGCCAUUCAGCAAUUUCGA__CUGGCCAAGUAUUUAAGCCAUUUGCACUAAUGAAUGCGGUCGCGCAUUUUUAAGCGCCC________AC__AAU ....................(((((.(((((.......(((...((((...........)))).)))......)))))))))).((((........))))................. (-16.12 = -15.99 + -0.13)

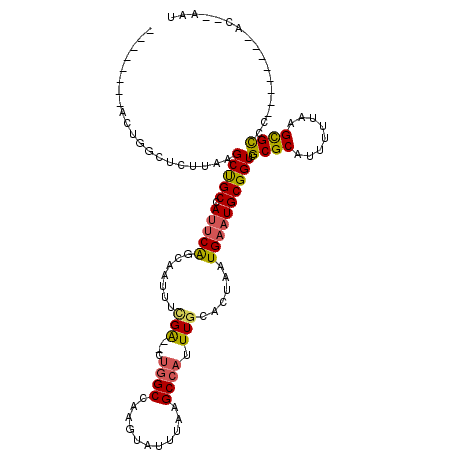
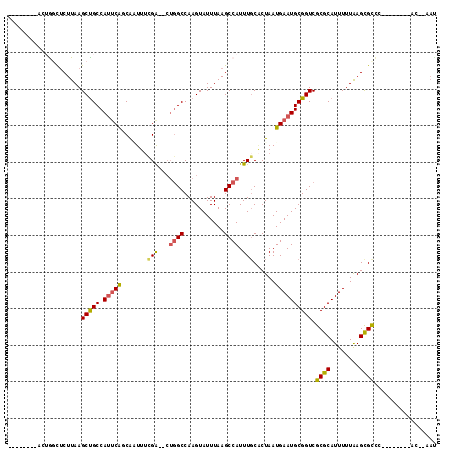
| Location | 12,342,079 – 12,342,176 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.45 |
| Mean single sequence MFE | -28.83 |
| Consensus MFE | -14.66 |
| Energy contribution | -15.25 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12342079 97 - 22407834 GUUUUGU--------GGGCGCUUAAAAAUGCGCGACCGCAUUCAUUAGAGAAAAUGGCUUAAAUACUUGGCCAG--UCGAAACUGAUGAAUGGCGGCUUAAGAAACC---------- (((((..--------((((((........))))..(((((((((((((.((...(((((.........))))).--))....))))))))).))))))...))))).---------- ( -34.40) >DroVir_CAF1 206104 93 - 1 ---UUGG--------GUACGCUUAAAAAUGCGCGACCGCAUUCAUUAGCGCAAAUGGCUUAAAUACUUGGC-----CCGCAAAUGUUGUGUGGCAGCUUAAGAACUAGU-------- ---...(--------((.(((........))((..((((((.((((.(((.....((((.........)))-----)))).))))..))))))..))....).)))...-------- ( -24.90) >DroPse_CAF1 240375 96 - 1 AUU-------------GGCGCUUAAAAAUGCGCGACCGCAUUCAUUAGUGCAAAUGGCUUAAAUACUUGGCCAG--UCGAAAUUGCUGAAUGGCAGCUUAAGAGCCAGUCU------ (((-------------(((.(((((.(((...((((.((((......))))...(((((.........))))))--)))..)))((((.....))))))))).))))))..------ ( -31.10) >DroWil_CAF1 206692 101 - 1 AUU--UU--------GAACGCUUAAAAAUGCGCGACCGCAUUCAUUAGUGCAAAUGGCUUAAAUACUUGGCCAGAACCAAAAUCGCUGAAUGGCAGCUUAAGUAGUAGAAG------ (((--((--------(..((((....((((((....))))))....))))....(((((.........)))))....)))))).((((.....))))..............------ ( -21.90) >DroAna_CAF1 152009 106 - 1 GUUUCGUAGGCAAUGGGGCACUCAAAAAUGCGCGACCGCAUUCAUUAGUGCAAAUGGCUUAAAUACUUGGCCAG--UCGAAAGUGGCGAAUGGCAGCUUAAGAGCCCC--------- ..............(((((.............((((.((((......))))...(((((.........))))))--))).((((.((.....)).))))....)))))--------- ( -29.60) >DroPer_CAF1 246690 102 - 1 AUU-------------GGCGCUUAAAAAUGCGCGACCGCAUUCAUUAGUGCAAAUGGCUUAAAUACUUGGCCAG--UCGAAAUUGCUGAAUGGCAGCUUAAGAGCCAGUCCCCUUAC (((-------------(((.(((((.(((...((((.((((......))))...(((((.........))))))--)))..)))((((.....))))))))).))))))........ ( -31.10) >consensus AUU__GU________GGGCGCUUAAAAAUGCGCGACCGCAUUCAUUAGUGCAAAUGGCUUAAAUACUUGGCCAG__UCGAAAUUGCUGAAUGGCAGCUUAAGAACCAGU________ ..................((((....((((((....))))))....))))....(((((.........)))))...........((((.....)))).................... (-14.66 = -15.25 + 0.59)

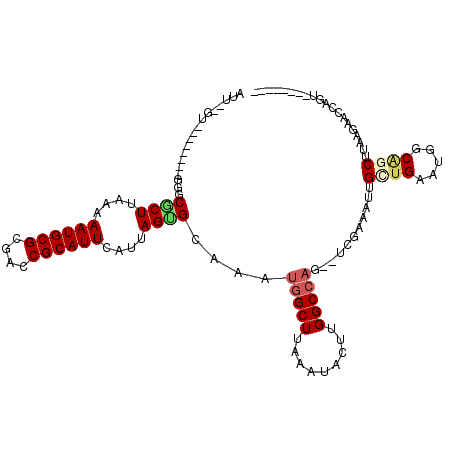
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:53 2006