| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,333,585 – 12,333,685 |
| Length | 100 |
| Max. P | 0.676411 |

| Location | 12,333,585 – 12,333,685 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.77 |
| Mean single sequence MFE | -34.03 |
| Consensus MFE | -21.44 |
| Energy contribution | -21.92 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.676411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
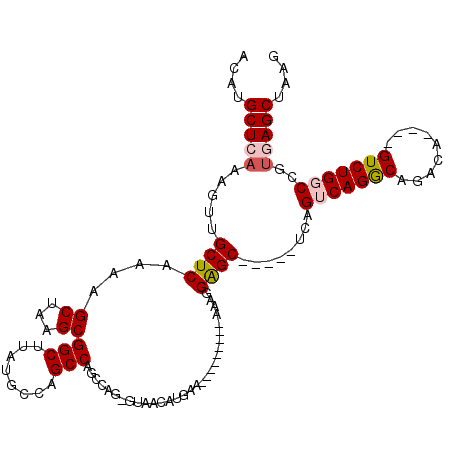
>2L_DroMel_CAF1 12333585 100 + 22407834 ACAUGCUCAAAGUUGCUCAAAAGCUAAGCGGCUUAUGCCAGCCAGCCAG-GUAACAUGAA---------AAAGGAGC-----UCAGUCAGGCAGACA----GUCUGGCCGUGAGCUAAG ...........((((((.....(((..(((((....))).)).)))..)-))))).....---------.....(((-----((((((((((.....----)))))))..))))))... ( -31.20) >DroPse_CAF1 230480 104 + 1 ACAUGCUCAAAGUUGCUCAAAAGCUAAGCGGCUUAUGACUGCCAUGGGG---------------GGGGCAGAGGGGCACAAUUCAGUCAGGCAGACACACUGUCUGCCAGUCAGCUAAG ...(((((.((((((((.........))))))))....(((((......---------------..)))))..)))))...........(((((((.....)))))))........... ( -35.60) >DroSim_CAF1 161630 100 + 1 ACAUGCUCAAAGUUGCUCAAAAGCUAAGCGGCUUAUGCCAGCCAGCCAU-GUAACAUGAA---------AAAGGAGC-----UCAGUCAGGCAGACA----GUCUGGCCGUGAGCUAAG ....(((((.....((((....(((..(((((....))).)).)))(((-(...))))..---------....))))-----...(((((((.....----)))))))..))))).... ( -31.90) >DroEre_CAF1 156132 100 + 1 ACAUGCUCAAAGUUGCUCAAAAGCUAAGCGGCUUAUGCCAGCCAGCCAG-GUAACAUGAA---------AAAGGAGC-----UCAGUCAGGCAGACA----GUCUGGCCGAGAGCUAAG ....((((...((((((.....(((..(((((....))).)).)))..)-))))).....---------........-----((.(((((((.....----))))))).)))))).... ( -30.60) >DroYak_CAF1 162831 100 + 1 ACAUGCUCAAAGUUGCUCAAAAGCUAAGCGGCUUAUGCCAGCCAGCCAG-GUAACAUGAA---------AAAGGAGC-----UCAGUCAGACAGACA----GUCUGGCCGUGAGCUAAG ...........((((((.....(((..(((((....))).)).)))..)-))))).....---------.....(((-----((((((((((.....----)))))))..))))))... ( -31.80) >DroPer_CAF1 235944 119 + 1 ACAUGCUCAAAGUUGCUCAAAAGCUAAGCGGCUUAUGCCUGCCACGGGGUGGGGCAUGAAGGGGUGGGCAGAGGGGCACAAUUCAGUCAGGCAGACACACUGUCUGCCAGUCAGCUAAG ...(((((....(((((((...(((....)))(((((((..((....))..).)))))).....)))))))..)))))...........(((((((.....)))))))........... ( -43.10) >consensus ACAUGCUCAAAGUUGCUCAAAAGCUAAGCGGCUUAUGCCAGCCAGCCAG_GUAACAUGAA_________AAAGGAGC_____UCAGUCAGGCAGACA____GUCUGGCCGUGAGCUAAG ....(((((.....((((....((...))(((........)))..............................))))........(((((((.........)))))))..))))).... (-21.44 = -21.92 + 0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:49 2006