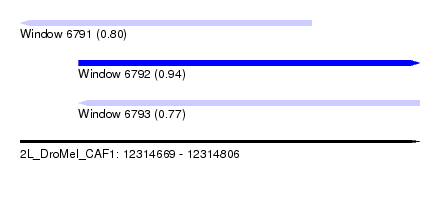
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,314,669 – 12,314,806 |
| Length | 137 |
| Max. P | 0.936730 |

| Location | 12,314,669 – 12,314,769 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.52 |
| Mean single sequence MFE | -18.02 |
| Consensus MFE | -12.40 |
| Energy contribution | -12.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.803992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12314669 100 - 22407834 UAAUCUUAUUAGCAGUUUGUUGUUCGAACCUAAUCAAUUACCAACGUAAUUACAAACUUUGCUAAUCUGACUCGGUUUCCCCCCAGA-AACGCUUUUCCAG------------------- ...((..(((((((((((((.....((......))((((((....))))))))))))..)))))))..))....(((((......))-)))..........------------------- ( -18.10) >DroVir_CAF1 168602 99 - 1 UAAUCUUAUUAGCAGUUUGUUGUUCGAACCUAAUCAAUUACCAACGUAAUUACAAACUUUGCUAAUCUGCUUAGCUCACA--GCGCA-CACA------G-A--GCGAAA---------GG ........((((((((((((.....((......))((((((....))))))))))))..))))))((((....((.....--))...-..))------)-)--.(....---------). ( -17.90) >DroPse_CAF1 203714 109 - 1 UAAUCUUAUUAGCAGUUUGUUGUUCGAACCUAAUCAAUUACCAACGUAAUUACAAACUUUGCUAAUCUGCCUCGGUCUCC--CCCGACAGCGCUACCCAAAAACCAAAA---------GA ...(((((((((((((((((.....((......))((((((....))))))))))))..)))))))..((.((((.....--.))))..))................))---------)) ( -19.30) >DroGri_CAF1 177291 108 - 1 UAAUCUUAUUAGCAGUUUGUUGUUCGAACCUAAUCAAUUACCAACGUAAUUACAAACUUUGCUAAUCUGCUUAGGUCGCA--ACAGA-UUCA------C-A--UCGAACAGGGCACAGAG ..............((.(.((((((((....((((((((((....)))))).....((..((......))..))......--...))-))..------.-.--)))))))).).)).... ( -19.60) >DroAna_CAF1 126109 98 - 1 UAAUCUUAUUAGCAGUUUGUUGUUCGAACCUAAUCAAUUACCAACGUAAUUACAAACUUUGCUAAUCUCCCUCGGUUUCC-CCCAGC-AACGAAUUUCC-G------------------- .......(((((((((((((.....((......))((((((....))))))))))))..)))))))......(((.(((.-......-...)))...))-)------------------- ( -13.90) >DroPer_CAF1 209195 109 - 1 UAAUCUUAUUAGCAGUUUGUUGUUCGAACCUAAUCAAUUACCAACGUAAUUACAAACUUUGCUAAUCUGCCUCGGUCUCC--CCCGACAGCGCUACCCAAAAACCAAAA---------GA ...(((((((((((((((((.....((......))((((((....))))))))))))..)))))))..((.((((.....--.))))..))................))---------)) ( -19.30) >consensus UAAUCUUAUUAGCAGUUUGUUGUUCGAACCUAAUCAAUUACCAACGUAAUUACAAACUUUGCUAAUCUGCCUCGGUCUCC__CCAGA_AACGCU___CC_A___C_AAA_________G_ .......(((((((((((((.....((......))((((((....))))))))))))..)))))))...................................................... (-12.40 = -12.40 + 0.00)

| Location | 12,314,689 – 12,314,806 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.21 |
| Mean single sequence MFE | -24.75 |
| Consensus MFE | -22.42 |
| Energy contribution | -22.62 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.936730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12314689 117 + 22407834 GGAAACCGAGUCAGAUUAGCAAAGUUUGUAAUUACGUUGGUAAUUGAUUAGGUUCGAACAACAAACUGCUAAUAAGAUUAUUGCAGUUGAGAACAAACCGAGCUGAAAAAACAAA---AG (....).........(((((...((((((......((((....((((......)))).)))).((((((.((((....))))))))))....))))))...))))).........---.. ( -23.80) >DroVir_CAF1 168621 119 + 1 UGUGAGCUAAGCAGAUUAGCAAAGUUUGUAAUUACGUUGGUAAUUGAUUAGGUUCGAACAACAAACUGCUAAUAAGAUUAUUGCAGUUGAGAACAAACCGAGCUGAAAAAACAACU-CAA .....(((((.....)))))...((((((......((((....((((......)))).)))).((((((.((((....))))))))))....)))))).(((.((......)).))-).. ( -26.10) >DroGri_CAF1 177319 119 + 1 UGCGACCUAAGCAGAUUAGCAAAGUUUGUAAUUACGUUGGUAAUUGAUUAGGUUCGAACAACAAACUGCUAAUAAGAUUAUUGCAGUCGAGAACAAACCGAGCUGAAAAAACAACU-CAA ..((((((((...(((((.(((.((........)).))).)))))..))))).)))........(((((.((((....)))))))))............(((.((......)).))-).. ( -24.70) >DroEre_CAF1 135206 117 + 1 GGAAACAGCGCCAGAUUAGCAAAGUUUGUAAUUACGUUGGUAAUUGAUUAGGUUCGAACAACAAACUGCUAAUAAGAUUAUUGCAGUUGAGAACAAACCGAGCUGAAAAAACAAA---AG .....((((((.......))...((((((......((((....((((......)))).)))).((((((.((((....))))))))))....))))))...))))..........---.. ( -24.80) >DroMoj_CAF1 179368 120 + 1 UGGGCGCUAAGCAGAUUAGCAAAGUUUGUAAUUACGUUGGUAAUUGAUUAGGUUCGAACAACAAACUGCUAAUAAGAUUAUUGCAGUCGAGAGCAAACCGAGCUGAAAAAACAACUUCAG (.((((((((.....)))))...((((((......((((....((((......)))).))))..(((((.((((....))))))))).....))))))...))).).............. ( -25.30) >DroAna_CAF1 126127 117 + 1 GGAAACCGAGGGAGAUUAGCAAAGUUUGUAAUUACGUUGGUAAUUGAUUAGGUUCGAACAACAAACUGCUAAUAAGAUUAUUGCAGUUGAGAACAAACCGAGCUGAAAAAACAAA---AA (....).........(((((...((((((......((((....((((......)))).)))).((((((.((((....))))))))))....))))))...))))).........---.. ( -23.80) >consensus GGAAACCGAAGCAGAUUAGCAAAGUUUGUAAUUACGUUGGUAAUUGAUUAGGUUCGAACAACAAACUGCUAAUAAGAUUAUUGCAGUUGAGAACAAACCGAGCUGAAAAAACAAA___AA ...............(((((...((((((......((((....((((......)))).)))).((((((.((((....))))))))))....))))))...))))).............. (-22.42 = -22.62 + 0.19)

| Location | 12,314,689 – 12,314,806 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.21 |
| Mean single sequence MFE | -24.63 |
| Consensus MFE | -20.12 |
| Energy contribution | -19.90 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
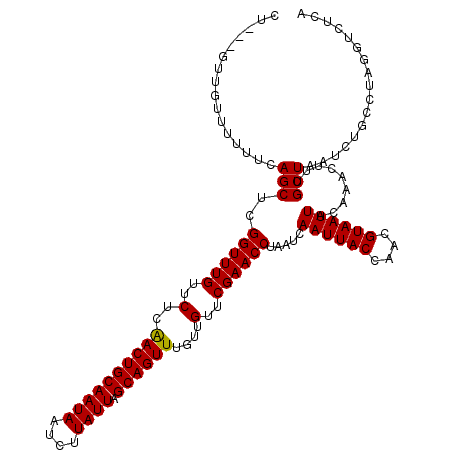
>2L_DroMel_CAF1 12314689 117 - 22407834 CU---UUUGUUUUUUCAGCUCGGUUUGUUCUCAACUGCAAUAAUCUUAUUAGCAGUUUGUUGUUCGAACCUAAUCAAUUACCAACGUAAUUACAAACUUUGCUAAUCUGACUCGGUUUCC ..---...(((.....(((..((((((..(.(((((((((((....)))).)))).)))..)..)))))).....((((((....)))))).........))).....)))......... ( -21.00) >DroVir_CAF1 168621 119 - 1 UUG-AGUUGUUUUUUCAGCUCGGUUUGUUCUCAACUGCAAUAAUCUUAUUAGCAGUUUGUUGUUCGAACCUAAUCAAUUACCAACGUAAUUACAAACUUUGCUAAUCUGCUUAGCUCACA ..(-(((((......))))))((((((..(.(((((((((((....)))).)))).)))..)..)))))).....((((((....)))))).........(((((.....)))))..... ( -28.50) >DroGri_CAF1 177319 119 - 1 UUG-AGUUGUUUUUUCAGCUCGGUUUGUUCUCGACUGCAAUAAUCUUAUUAGCAGUUUGUUGUUCGAACCUAAUCAAUUACCAACGUAAUUACAAACUUUGCUAAUCUGCUUAGGUCGCA ..(-(((((......))))))((((((..(..((((((((((....)))).))))))....)..)))))).....((((((....)))))).....((..((......))..))...... ( -28.80) >DroEre_CAF1 135206 117 - 1 CU---UUUGUUUUUUCAGCUCGGUUUGUUCUCAACUGCAAUAAUCUUAUUAGCAGUUUGUUGUUCGAACCUAAUCAAUUACCAACGUAAUUACAAACUUUGCUAAUCUGGCGCUGUUUCC ..---..........((((..((((((..(.(((((((((((....)))).)))).)))..)..)))))).....((((((....)))))).........(((.....)))))))..... ( -24.30) >DroMoj_CAF1 179368 120 - 1 CUGAAGUUGUUUUUUCAGCUCGGUUUGCUCUCGACUGCAAUAAUCUUAUUAGCAGUUUGUUGUUCGAACCUAAUCAAUUACCAACGUAAUUACAAACUUUGCUAAUCUGCUUAGCGCCCA ((((((......))))))...((((((..(..((((((((((....)))).))))))....)..)))))).....((((((....)))))).........(((((.....)))))..... ( -25.20) >DroAna_CAF1 126127 117 - 1 UU---UUUGUUUUUUCAGCUCGGUUUGUUCUCAACUGCAAUAAUCUUAUUAGCAGUUUGUUGUUCGAACCUAAUCAAUUACCAACGUAAUUACAAACUUUGCUAAUCUCCCUCGGUUUCC ..---...........(((..((((((..(.(((((((((((....)))).)))).)))..)..)))))).....((((((....)))))).........)))................. ( -20.00) >consensus CU___GUUGUUUUUUCAGCUCGGUUUGUUCUCAACUGCAAUAAUCUUAUUAGCAGUUUGUUGUUCGAACCUAAUCAAUUACCAACGUAAUUACAAACUUUGCUAAUCUGCCUAGGUCUCA ................(((..((((((..(..((((((((((....)))).))))))....)..)))))).....((((((....)))))).........)))................. (-20.12 = -19.90 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:44 2006