| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,308,602 – 12,308,700 |
| Length | 98 |
| Max. P | 0.566580 |

| Location | 12,308,602 – 12,308,700 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 80.45 |
| Mean single sequence MFE | -18.43 |
| Consensus MFE | -11.76 |
| Energy contribution | -13.85 |
| Covariance contribution | 2.09 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
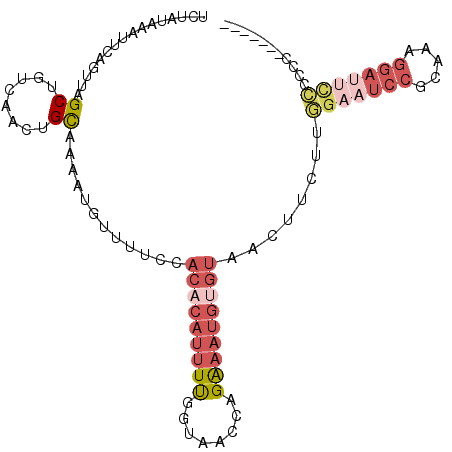
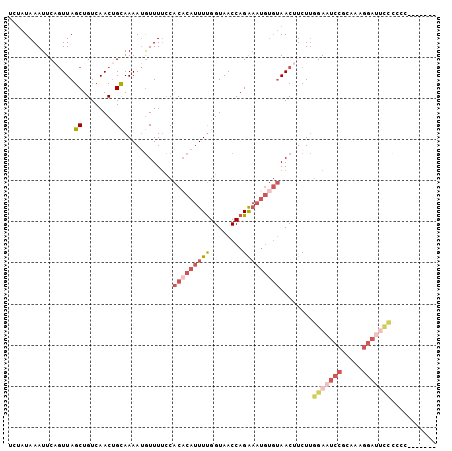
>2L_DroMel_CAF1 12308602 98 - 22407834 UCUAUAAAUUCAGUUAGCUGUCAACUGCAAAAUGUUUUCCACUCAUUUUGGUAACCAGAAAUGUGUAACUUCUUGGAAUCCGCAAAGGAUUCCCACCC------ ..........(((((.......)))))(((((((.........)))))))((.((((....)).)).)).....(((((((.....))))))).....------ ( -20.60) >DroSec_CAF1 131133 97 - 1 UCUAUAAAUUCAGUUAGCUGUCAACUGCAAAAUGUUUUCCACACAUUUUGGUAACCAGAAAUGUGUAACUUCUUGGAAUCCGCAAAGGAUUCCCGCC------- ..........(((....)))......((............(((((((((((...)))).)))))))........(((((((.....))))))).)).------- ( -22.80) >DroSim_CAF1 134835 97 - 1 UCUAUAAAUUCCGUUAGCUGUCAACUGCAAAAUGUUUUCCACACAUUUUGGUAACCAGAAAUGUGUAACUUCUUGGAAUCCGCAAAGGAUUCCCGCC------- ............(((.((........))............(((((((((((...)))).)))))))))).....(((((((.....)))))))....------- ( -21.80) >DroEre_CAF1 129046 86 - 1 UCUAUAAAUUCAGUUAGCUGUCAACUGCAAAAUGUUUUCCACACAUUUUGGUAACCAGAAAUGUGUAACUUCUCGGAAUCCUC------------------CAC ...........((((.((........))............(((((((((((...)))).)))))))))))....(((....))------------------).. ( -14.10) >DroYak_CAF1 135610 104 - 1 UCUAUAAAUUCAGUUAGCUGUCAACUGCAAAAUGUUUUCCACACAUUUUGGUAACCAGAAAUGUGUAACUUCUUGGAAUCCGCAAAGGAUUCCCCCCCGUUCGC ...........((((.((........))............(((((((((((...)))).)))))))))))....(((((((.....)))))))........... ( -22.30) >DroAna_CAF1 119746 85 - 1 ---CUAAAUUCUCUUAGCUGUCAACCGUAAAACAUUUCUCGCACAUUUCGGUAACCAGG-------AACUUCUUUGAAUCCACCAAGGACCUAUC--------- ---..............(((...((((.....................))))...))).-------....((((((.......))))))......--------- ( -9.00) >consensus UCUAUAAAUUCAGUUAGCUGUCAACUGCAAAAUGUUUUCCACACAUUUUGGUAACCAGAAAUGUGUAACUUCUUGGAAUCCGCAAAGGAUUCCCCCC_______ ................((........))............(((((((((........)))))))))........(((((((.....)))))))........... (-11.76 = -13.85 + 2.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:41 2006