| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,305,769 – 12,305,859 |
| Length | 90 |
| Max. P | 0.892078 |

| Location | 12,305,769 – 12,305,859 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 76.27 |
| Mean single sequence MFE | -18.20 |
| Consensus MFE | -9.60 |
| Energy contribution | -10.40 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12305769 90 + 22407834 -CCAAUGCCUGGGAGAGAUGUAGCAAAAAUCAAUAAGAUCAAUCAUCUUGUAUGCUCCUCUUUUGCUCCUCUUGCUCCCCUUUUGCUCCCC -.....((..((((((((.(.(((((((..(((((((((.....))))))).))......)))))))).)))..))))).....))..... ( -18.60) >DroSec_CAF1 128425 79 + 1 -CCAAUGCCUGGGAGAGAUGUAGCAAAAAUCAAUAAGAUCAAUCAUCUUGUAUGCUCC-----------UCCUGCUCCUCAUCUGCUCCCC -.........((((((((((.((((.....(((((((((.....))))))).))....-----------...))))...))))).))))). ( -21.30) >DroSim_CAF1 132093 79 + 1 -CCAAUGCCUGGGAGAGAUGUAGCAAAAAUCAAUAAGAUCAAUCAUCUUGUAUGCUCC-----------UCCUGCUCCUCAUCUGCUCCCC -.........((((((((((.((((.....(((((((((.....))))))).))....-----------...))))...))))).))))). ( -21.30) >DroYak_CAF1 132951 86 + 1 -CCAAUGCCUGGGAGAGAUGUAGCAAAAAUCAAUAAGAUCAAUCAUCCUGCAUGCUGCUCCACUGCUCCAC-UGC---UCCACUGCUCCCC -.........(((((((.((.((((...(((.....))).............((..((......))..)).-)))---).)))).))))). ( -16.80) >DroAna_CAF1 117277 82 + 1 GCAAAUGCCCAGGAGAGACGGACCAAAAAUCAAUAAGAUCAAUCAUCUUGUAUAUUUCACUUUGGCUACAU--------ACGCUGCUCCC- ((....))...((((((.((..((((((((..(((((((.....)))))))..)))....)))))......--------.)))).)))).- ( -13.00) >consensus _CCAAUGCCUGGGAGAGAUGUAGCAAAAAUCAAUAAGAUCAAUCAUCUUGUAUGCUCC_C____GCU_CUC_UGCUCCUCAUCUGCUCCCC ..........(((((.(((.........))).(((((((.....)))))))..................................))))). ( -9.60 = -10.40 + 0.80)

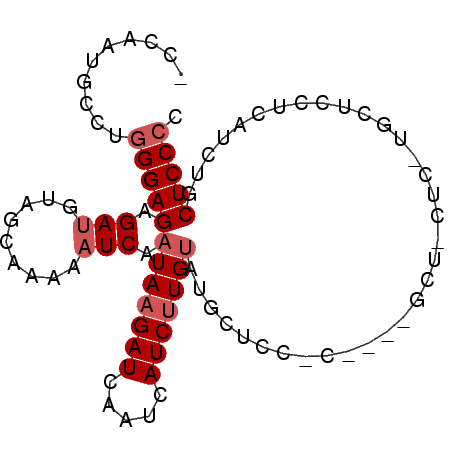

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:38 2006