| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,305,196 – 12,305,294 |
| Length | 98 |
| Max. P | 0.775730 |

| Location | 12,305,196 – 12,305,294 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 82.92 |
| Mean single sequence MFE | -24.83 |
| Consensus MFE | -18.49 |
| Energy contribution | -18.63 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
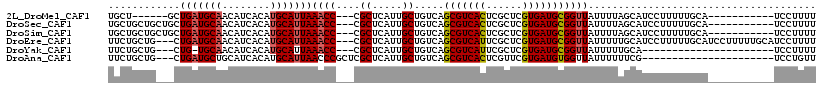
>2L_DroMel_CAF1 12305196 98 + 22407834 UGCU------GCUGAUGCAACAUCACAUGCAUUAAACC---CGCUCAUUGCUGUCAGCGUCACUCGCUCGUGAUGCGGUUAUUUUAGCAUCCUUUUUGCA-----------UCCUUUU ((((------(.(((((((........)))))))((((---.((.....)).....(((((((......)))))))))))....)))))...........-----------....... ( -25.90) >DroSec_CAF1 127853 104 + 1 UGCUGCUGCUGCUGAUGCAACAUCACAUGCAUUAAACC---CGCUCAUUGCUGUCAGCGUCACUCGCUCGUGAUGCGGUUAUUUUAGCAUCCUUUUUGCA-----------UCCUUUU .((....))....(((((((......((((....((((---.((.....)).....(((((((......)))))))))))......)))).....)))))-----------))..... ( -26.70) >DroSim_CAF1 131521 104 + 1 UGCUGCUGCUGCUGAUGCAACAUCACAUGCAUUAAACC---CGCUCAUUGCUGUCAGCGUCACUCGCUCGUGAUGCGGUUAUUUUAGCAUCCUUUUUGCA-----------UCCUUUU .((....))....(((((((......((((....((((---.((.....)).....(((((((......)))))))))))......)))).....)))))-----------))..... ( -26.70) >DroEre_CAF1 125857 112 + 1 UUCUGCUG---CUGAUGCAACAUCACAUGCAUUAAACC---CGCUCAUUGCUGUCAGCGUCAUUCGCUCGUGAUGCGGUUAUUUUUGCAUCCUUUUUGCAUCCUUUUUGCAUCCUUUU ...(((..---..(((((((......(((((...((((---.((.....)).....(((((((......))))))))))).....))))).....)))))))......)))....... ( -28.00) >DroYak_CAF1 132428 89 + 1 UUCUGCUG---CUG-UGCAACAUCACAUGCAUUAAACC---CGCUCAUUGCUGUCAGCGUCAUUCGCUCGUGAUGCGGUUAUUUUUGCA----------------------UCCUUUU ...(((((---(((-((......)))).)))...((((---.((.....)).....(((((((......)))))))))))......)))----------------------....... ( -21.20) >DroAna_CAF1 116740 93 + 1 UUCUGCUG---CUGAUGCUGCAUCACAUGCAUUAACCCGCUCGCUCAUUGCUGUCAGCGUCACUCGUUCGUGAUGUGGUUAUUUUUUCG----------------------UCCUGUU ...(((((---.(((((...))))))).))).(((((.....((.....)).....(((((((......))))))))))))........----------------------....... ( -20.50) >consensus UGCUGCUG___CUGAUGCAACAUCACAUGCAUUAAACC___CGCUCAUUGCUGUCAGCGUCACUCGCUCGUGAUGCGGUUAUUUUAGCAUCCUUUUUGCA___________UCCUUUU ............(((((((........)))))))((((....((.....)).....(((((((......)))))))))))...................................... (-18.49 = -18.63 + 0.14)
| Location | 12,305,196 – 12,305,294 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.92 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -19.42 |
| Energy contribution | -20.80 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
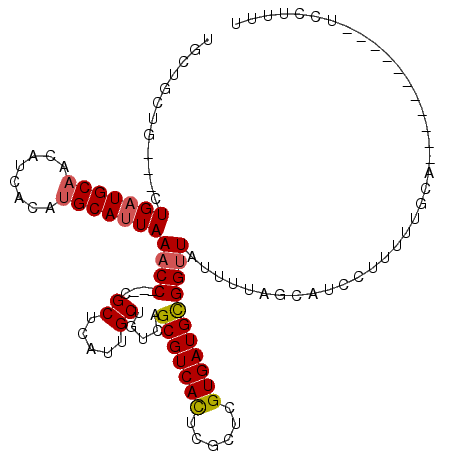
>2L_DroMel_CAF1 12305196 98 - 22407834 AAAAGGA-----------UGCAAAAAGGAUGCUAAAAUAACCGCAUCACGAGCGAGUGACGCUGACAGCAAUGAGCG---GGUUUAAUGCAUGUGAUGUUGCAUCAGC------AGCA .....((-----------(((((...(.((((........((((.((((......)))).((.....)).....)))---).......)))).)....)))))))...------.... ( -28.86) >DroSec_CAF1 127853 104 - 1 AAAAGGA-----------UGCAAAAAGGAUGCUAAAAUAACCGCAUCACGAGCGAGUGACGCUGACAGCAAUGAGCG---GGUUUAAUGCAUGUGAUGUUGCAUCAGCAGCAGCAGCA .....((-----------(((((...(.((((........((((.((((......)))).((.....)).....)))---).......)))).)....))))))).((....)).... ( -30.06) >DroSim_CAF1 131521 104 - 1 AAAAGGA-----------UGCAAAAAGGAUGCUAAAAUAACCGCAUCACGAGCGAGUGACGCUGACAGCAAUGAGCG---GGUUUAAUGCAUGUGAUGUUGCAUCAGCAGCAGCAGCA .....((-----------(((((...(.((((........((((.((((......)))).((.....)).....)))---).......)))).)....))))))).((....)).... ( -30.06) >DroEre_CAF1 125857 112 - 1 AAAAGGAUGCAAAAAGGAUGCAAAAAGGAUGCAAAAAUAACCGCAUCACGAGCGAAUGACGCUGACAGCAAUGAGCG---GGUUUAAUGCAUGUGAUGUUGCAUCAG---CAGCAGAA .......(((......(((((((...(.(((((.......((((......((((.....))))..((....)).)))---)......))))).)....)))))))..---..)))... ( -31.12) >DroYak_CAF1 132428 89 - 1 AAAAGGA----------------------UGCAAAAAUAACCGCAUCACGAGCGAAUGACGCUGACAGCAAUGAGCG---GGUUUAAUGCAUGUGAUGUUGCA-CAG---CAGCAGAA ....(.(----------------------((((.......((((......((((.....))))..((....)).)))---)......))))).)..((((((.-..)---)))))... ( -23.42) >DroAna_CAF1 116740 93 - 1 AACAGGA----------------------CGAAAAAAUAACCACAUCACGAACGAGUGACGCUGACAGCAAUGAGCGAGCGGGUUAAUGCAUGUGAUGCAGCAUCAG---CAGCAGAA .......----------------------........(((((...((((......))))((((....((.....)).))))))))).(((.(((((((...))))).---)))))... ( -22.20) >consensus AAAAGGA___________UGCAAAAAGGAUGCAAAAAUAACCGCAUCACGAGCGAGUGACGCUGACAGCAAUGAGCG___GGUUUAAUGCAUGUGAUGUUGCAUCAG___CAGCAGAA ...........................(((((..........((((((((((((.....))))....(((.(((((.....))))).))).)))))))).)))))............. (-19.42 = -20.80 + 1.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:37 2006