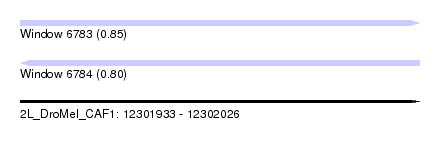
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,301,933 – 12,302,026 |
| Length | 93 |
| Max. P | 0.845785 |

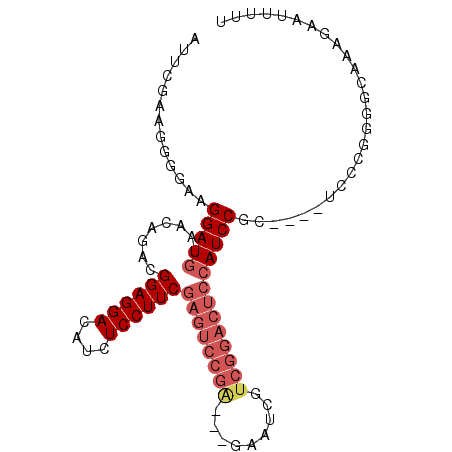
| Location | 12,301,933 – 12,302,026 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 83.30 |
| Mean single sequence MFE | -30.78 |
| Consensus MFE | -20.91 |
| Energy contribution | -22.59 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12301933 93 + 22407834 AAAAAUUCUUUGCCCCGGGAGCGAGCGGAUGGAGUCCGACGAU-----UCGGACUCGAAGGAGAUGUCCUCCGUCUGUUACAUCCUUUCACUUCGAAU .................((((.(((.(((((((((((((....-----))))))))((.((((.....)))).)).....))))).))).)))).... ( -35.00) >DroSec_CAF1 124655 93 + 1 AAAAAUUCUUUGCCCCGGCA----GCGGAUGGAGUCCGACGAUUCAU-UCGGACUCGAAGGAGAUGUCCUCCGUCUGUUACAUCCUUCCCCUGCGAAU ....((((.((((....)))----).(((((((((((((........-))))))))((.((((.....)))).)).....))))).........)))) ( -32.30) >DroSim_CAF1 128312 93 + 1 AAAAAUUCUUUGCCCCGGCA----GCGGAUGGAGUCCGACGAUUCAU-UCGGACUCGAAGGAGAUGUCCUCCGUCUGUUACAUCCUUCCCCUGCGAAU ....((((.((((....)))----).(((((((((((((........-))))))))((.((((.....)))).)).....))))).........)))) ( -32.30) >DroEre_CAF1 122691 75 + 1 AAAAAUUCUCUGCC-CGGGA----GCGGAU------------------UCGGACUCGAAGGAGAUGUCCUCCGUCUGUUACAUCCUGCGCCUUGGAGU .......((((((.-(((((----((((((------------------..((((((......)).))))...))))))....))))).))...)))). ( -23.30) >DroYak_CAF1 129090 94 + 1 AAAAAUUCUUUGCCCCAGGA----GCGGAUGGAGUCCGACGAUUCGGACCGGACUCGAAGGAGAUGUCCUCCGUCUGUUACAUCCUUUCCCUUCGAAC ................((((----(((((((((((((((....)))))).((((((......)).)))))))))))))....))))............ ( -31.00) >consensus AAAAAUUCUUUGCCCCGGGA____GCGGAUGGAGUCCGACGAUUC___UCGGACUCGAAGGAGAUGUCCUCCGUCUGUUACAUCCUUCCCCUUCGAAU ....((((........((((....((((((.(((((((...........)))))))...((((.....))))))))))....))))........)))) (-20.91 = -22.59 + 1.68)

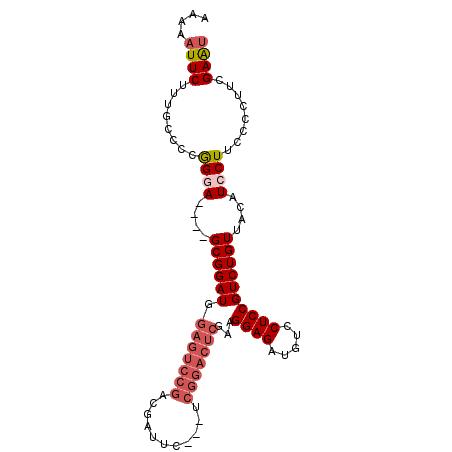
| Location | 12,301,933 – 12,302,026 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 83.30 |
| Mean single sequence MFE | -32.21 |
| Consensus MFE | -19.96 |
| Energy contribution | -21.64 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12301933 93 - 22407834 AUUCGAAGUGAAAGGAUGUAACAGACGGAGGACAUCUCCUUCGAGUCCGA-----AUCGUCGGACUCCAUCCGCUCGCUCCCGGGGCAAAGAAUUUUU .((((.(((((..(((((........((((((....))))))((((((((-----....)))))))))))))..)))))..))))............. ( -35.40) >DroSec_CAF1 124655 93 - 1 AUUCGCAGGGGAAGGAUGUAACAGACGGAGGACAUCUCCUUCGAGUCCGA-AUGAAUCGUCGGACUCCAUCCGC----UGCCGGGGCAAAGAAUUUUU .(((((((.(((.(((.........(((((((....))))))).((((((-........))))))))).))).)----)).))))............. ( -34.40) >DroSim_CAF1 128312 93 - 1 AUUCGCAGGGGAAGGAUGUAACAGACGGAGGACAUCUCCUUCGAGUCCGA-AUGAAUCGUCGGACUCCAUCCGC----UGCCGGGGCAAAGAAUUUUU .(((((((.(((.(((.........(((((((....))))))).((((((-........))))))))).))).)----)).))))............. ( -34.40) >DroEre_CAF1 122691 75 - 1 ACUCCAAGGCGCAGGAUGUAACAGACGGAGGACAUCUCCUUCGAGUCCGA------------------AUCCGC----UCCCG-GGCAGAGAAUUUUU .((((..((.((.((((........(((((((....))))))).......------------------))))))----.))..-)).))......... ( -22.06) >DroYak_CAF1 129090 94 - 1 GUUCGAAGGGAAAGGAUGUAACAGACGGAGGACAUCUCCUUCGAGUCCGGUCCGAAUCGUCGGACUCCAUCCGC----UCCUGGGGCAAAGAAUUUUU ((((...((((..(((((........((((((....))))))((((((((.(......))))))))))))))..----)))).))))........... ( -34.80) >consensus AUUCGAAGGGGAAGGAUGUAACAGACGGAGGACAUCUCCUUCGAGUCCGA___GAAUCGUCGGACUCCAUCCGC____UCCCGGGGCAAAGAAUUUUU .............(((((........((((((....))))))((((((((.........))))))))))))).......................... (-19.96 = -21.64 + 1.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:35 2006