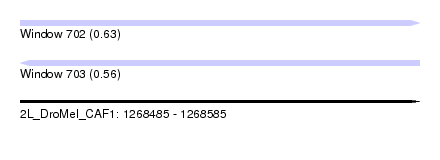
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,268,485 – 1,268,585 |
| Length | 100 |
| Max. P | 0.633171 |

| Location | 1,268,485 – 1,268,585 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 87.79 |
| Mean single sequence MFE | -38.57 |
| Consensus MFE | -29.03 |
| Energy contribution | -29.42 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
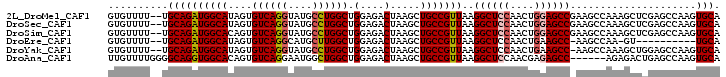
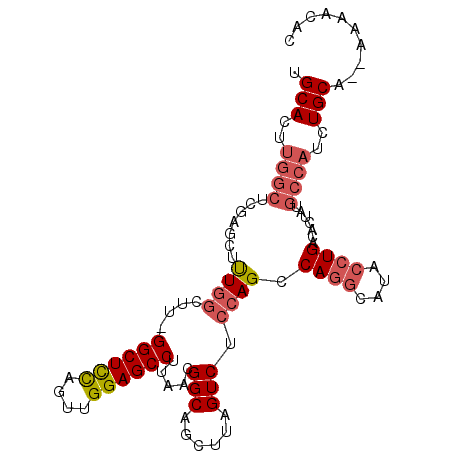
>2L_DroMel_CAF1 1268485 100 + 22407834 GUGUUUU--UGCAGAUGGCAUAGUGUCAGGUAUGCCUGGCUGGAGACUAAGCUGCCGUUAAGGCUCCAACUGGAGCCGAAGCCAAAGCUCGAGCCAAGUGCA .......--((((..((((.....((((((....)))))).(....)..((((...(((..((((((....))))))..)))...))))...))))..)))) ( -39.90) >DroSec_CAF1 28090 100 + 1 GUGUUUU--UGCAGAUGGCAUAGUGUCAGGUAUGCCUGGCUGGAGACUAAGCUGCCGUUAAGGCUCCAACUGGAGCCGAAGCCAAAGCUCGAGCCAAGUGCA .......--((((..((((.....((((((....)))))).(....)..((((...(((..((((((....))))))..)))...))))...))))..)))) ( -39.90) >DroSim_CAF1 29416 100 + 1 GUGUUUU--UGCAGAUGGCACAGUGUCAGGUAUGCCUGGCUGGAGACUAAGCUGCCGUUAAGGCUCCAACUGGAGCCGAAGCCAAAGCUCGAGCCAAGUGCA .......--((((..((((.....((((((....)))))).(....)..((((...(((..((((((....))))))..)))...))))...))))..)))) ( -39.90) >DroEre_CAF1 40687 88 + 1 GUGUUUU--UGCAGAUGGCAUAGUGUCAGGCAUGCUUGGCUGGAGACUAAGCUGCCGUUAAGGCUCCAACUGAAGCC-AAGCCAA-GU----------UGCA .......--(((((.((((.........((((.(((((((....).)))))))))).....((((.(....).))))-..)))).-.)----------)))) ( -30.90) >DroYak_CAF1 29148 99 + 1 GUGUUUU--UGCAGAUGGCAUAGUGUCAGGUAUGCCUGGCUGGAGACUAAGCUGCCGUUAAGGCUCCAACUGAAGCC-AAGCCAAAGCUGGAGCCAAGUGCA .......--(((((((((((....((((((....)))))).(....).....)))))))..(((((((.((...((.-..))...)).)))))))...)))) ( -38.10) >DroAna_CAF1 52337 96 + 1 UUGUUUUGGGGCAGGUGGCACAGUGUCAGGAAUGGCUGGCUGGAGACUAAGCUGCCGUUAAGGCUCCAACGAGAGCC------AGAGACUGAGCCAAGUGCA ..........(((..((((.((((.((...((((((.(((((....)..))))))))))..(((((......)))))------.)).)))).))))..))). ( -42.70) >consensus GUGUUUU__UGCAGAUGGCAUAGUGUCAGGUAUGCCUGGCUGGAGACUAAGCUGCCGUUAAGGCUCCAACUGGAGCC_AAGCCAAAGCUCGAGCCAAGUGCA ..........((((((((((....((((((....)))))).(....).....)))))))..((((((....)))))).....................))). (-29.03 = -29.42 + 0.39)
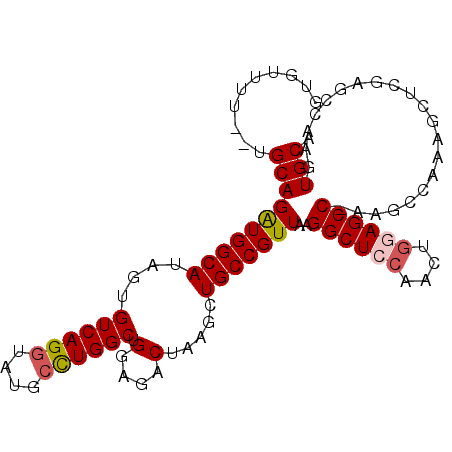
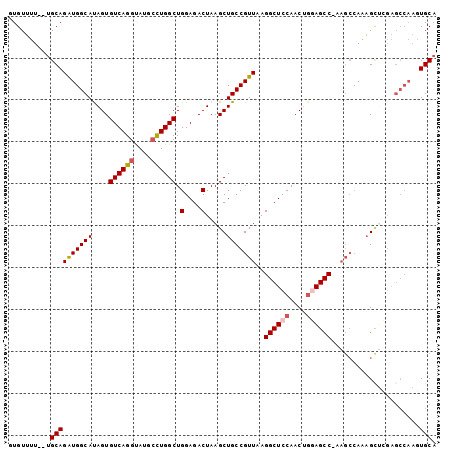
| Location | 1,268,485 – 1,268,585 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 87.79 |
| Mean single sequence MFE | -35.67 |
| Consensus MFE | -24.47 |
| Energy contribution | -25.30 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.55 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1268485 100 - 22407834 UGCACUUGGCUCGAGCUUUGGCUUCGGCUCCAGUUGGAGCCUUAACGGCAGCUUAGUCUCCAGCCAGGCAUACCUGACACUAUGCCAUCUGCA--AAAACAC ((((..(((((.(((((..((((((((((((....)))))).....)).)))).)).))).)))))((((((........))))))...))))--....... ( -38.20) >DroSec_CAF1 28090 100 - 1 UGCACUUGGCUCGAGCUUUGGCUUCGGCUCCAGUUGGAGCCUUAACGGCAGCUUAGUCUCCAGCCAGGCAUACCUGACACUAUGCCAUCUGCA--AAAACAC ((((..(((((.(((((..((((((((((((....)))))).....)).)))).)).))).)))))((((((........))))))...))))--....... ( -38.20) >DroSim_CAF1 29416 100 - 1 UGCACUUGGCUCGAGCUUUGGCUUCGGCUCCAGUUGGAGCCUUAACGGCAGCUUAGUCUCCAGCCAGGCAUACCUGACACUGUGCCAUCUGCA--AAAACAC ((((..(((((.(((((..((((((((((((....)))))).....)).)))).)).))).)))))((((((........))))))...))))--....... ( -38.20) >DroEre_CAF1 40687 88 - 1 UGCA----------AC-UUGGCUU-GGCUUCAGUUGGAGCCUUAACGGCAGCUUAGUCUCCAGCCAAGCAUGCCUGACACUAUGCCAUCUGCA--AAAACAC ((((----------..-.((((..-((((((....)))))).....((((((((.((.....)).)))).)))).........))))..))))--....... ( -30.30) >DroYak_CAF1 29148 99 - 1 UGCACUUGGCUCCAGCUUUGGCUU-GGCUUCAGUUGGAGCCUUAACGGCAGCUUAGUCUCCAGCCAGGCAUACCUGACACUAUGCCAUCUGCA--AAAACAC ((((...((((((((((..(((..-.)))..)))))))))).....(((((((........)))((((....))))......))))...))))--....... ( -36.50) >DroAna_CAF1 52337 96 - 1 UGCACUUGGCUCAGUCUCU------GGCUCUCGUUGGAGCCUUAACGGCAGCUUAGUCUCCAGCCAGCCAUUCCUGACACUGUGCCACCUGCCCCAAAACAA .(((..((((.((((.(((------((((...(((((((.((...(....)...)).))))))).))))).....)).)))).))))..))).......... ( -32.60) >consensus UGCACUUGGCUCGAGCUUUGGCUU_GGCUCCAGUUGGAGCCUUAACGGCAGCUUAGUCUCCAGCCAGGCAUACCUGACACUAUGCCAUCUGCA__AAAACAC .(((..((((.......((((....((((((....)))))).....(((......))).)))).((((....)))).......))))..))).......... (-24.47 = -25.30 + 0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:06 2006