
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,298,695 – 12,298,785 |
| Length | 90 |
| Max. P | 0.829039 |

| Location | 12,298,695 – 12,298,785 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 74.86 |
| Mean single sequence MFE | -19.47 |
| Consensus MFE | -15.09 |
| Energy contribution | -14.59 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.24 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829039 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12298695 90 + 22407834 UGUGCCUUCGUCACAUUCGCCACCAAGCACGCUGCCAUUUCGGCCAUUAAAGUGGUGAGUAGCGA------A--U-ACU--C-GUAUCUUGUAGAU--AAAUGU ......(((((...(((((((((..((....))(((.....))).......))))))))).))))------)--.-((.--.-.((((.....)))--)...)) ( -21.30) >DroVir_CAF1 147249 97 + 1 UGUGCCUUUGUUACCUUUGCCACCAAACACGCUGCCAUCUCGGCCAUUAAAGUGGUAAGUGGCUGGCUUUUAUAA-AC-----AUAUAAUAUAAAUACAAAUC- ((((((...(((((..(((((((.......((((......)))).......)))))))))))).))).((((((.-..-----......)))))).)))....- ( -20.74) >DroPse_CAF1 183963 93 + 1 UGUGCCUUUGUCACAUUCGCCACCAAGCACGCCGCCAUUUCGGCCAUUAAAGUGGUAAGUACUUG------AAGU-ACU--ACAUAUAUCCCCACA--AAAUCA ((((..............(((((.......((((......)))).......))))).(((((...------..))-)))--...........))))--...... ( -19.14) >DroEre_CAF1 119452 90 + 1 UGUGCCUUCGUCACAUUCGCUACCAAACACGCCGCCAUUUCGGCCAUUAAAGUGGUGAGUACUGA------A--C-ACU--C-UUAUCUUGUGGAU--ACAUGU ((((......(((.(((((((((.......((((......)))).......)))))))))..)))------.--(-((.--.-.......)))..)--)))... ( -19.04) >DroWil_CAF1 142669 87 + 1 UGUGCCUUUGUCACAUUUGCCACAAAACACGCUGCCAUUUCGGCCAUUAAAGUGGUAAGUAA--A------A--UCGCUGGC-UCAACUUA----A--AAAUGG ((.(((..((....(((((((((.......((((......)))).......)))))))))..--.------.--.))..)))-.)).....----.--...... ( -20.14) >DroYak_CAF1 125789 90 + 1 UGUGCCUUCGUCACAUUCGCCACCAAACACGCUGCCAUUUCGGCCAUUAAAGUGGUGAGUACUAA------A--U-GCU--C-UUCUUUUGUAAAU--AAAUAU .((((....).)))(((((((((.......((((......)))).......))))))))).....------.--.-...--.-.............--...... ( -16.44) >consensus UGUGCCUUCGUCACAUUCGCCACCAAACACGCUGCCAUUUCGGCCAUUAAAGUGGUAAGUACUGA______A__U_ACU__C_UUAUCUUGUAAAU__AAAUGU .((((....).)))(((((((((.......((((......)))).......)))))))))............................................ (-15.09 = -14.59 + -0.50)

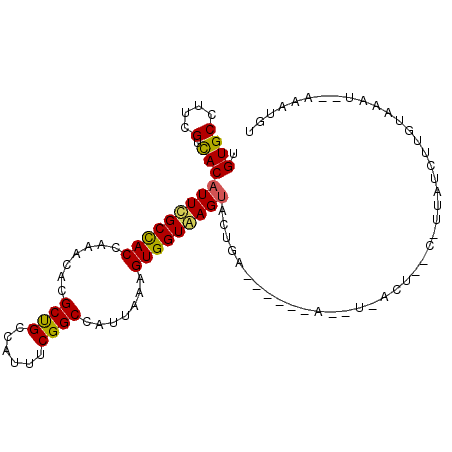
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:28 2006