| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,296,640 – 12,296,776 |
| Length | 136 |
| Max. P | 0.567655 |

| Location | 12,296,640 – 12,296,752 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.17 |
| Mean single sequence MFE | -30.79 |
| Consensus MFE | -19.74 |
| Energy contribution | -19.83 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.515793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
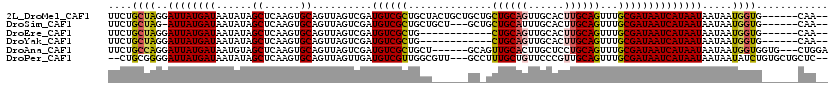
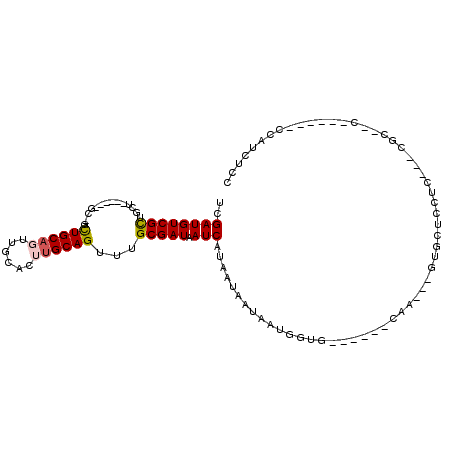
>2L_DroMel_CAF1 12296640 112 + 22407834 UUCUGCUAGGAUUAUGAUAAUAUAGCUCAAGUGCAGUUAGUCGAUGUCGCUGCUACUGCUGCUGCUGCAGUUGCACUUGCAGUUUGCGAUAAUCAUAAUAAUAAUGGUG------CAA-- ....((((..((((((((.((..((((((((((((((.((.((....)))))))...(((((....))))).))))))).))))....)).)))))))).....)))).------...-- ( -32.80) >DroSim_CAF1 123117 108 + 1 UUCUGCUAG-AUUAUGAUAAUAUAGCUCAAGUGCAGUUAGUCGAUGUCGCUGCUGCU---GCUGCUGCAUUUGCACUUGCAGUUUGCGAUAAUCAUAAUAAUAAUGGUG------CAA-- ....((((.-((((((((.((..((((((((((((.......(((((.((.((....---)).)).))))))))))))).))))....)).)))))))).....)))).------...-- ( -31.41) >DroEre_CAF1 117496 100 + 1 UUCUGCUAGGAUUAUGAUAAUAUAGCUCAAGUGCAGUUAGUCGAUGUCGCUG------------CUGCAGUUGCACUUGCAGUUUGCGAUAAUCAUAAUAAUAAUGGUG------CAA-- ....((((..((((((((......((......))..........((((((.(------------((((((......)))))))..)))))))))))))).....)))).------...-- ( -27.80) >DroYak_CAF1 123788 100 + 1 UUCUGCUAGGAUUAUGAUAAUAUAGCUCAAGUGCAGUUAGUCGAUGUCGCUG------------CUGCAGUUGCACUUGCAGUUUGCGAUAAUCAUAAUAAUAAUGGUG------CAA-- ....((((..((((((((......((......))..........((((((.(------------((((((......)))))))..)))))))))))))).....)))).------...-- ( -27.80) >DroAna_CAF1 108148 111 + 1 UUCUGCCAGGAUUAUGAUAAUGUAGCUCAAGUGCAGUUAGUCGAUGUCGCUGCU------GCAGUUGCACUUGCUCCUGCAGUUUGCGAUAAUCAUAAUAAUAAUGGUGGUG---CUGGA ..(..(((..((((((((......((......))..........((((((.(((------((((..((....))..)))))))..)))))))))))))).....)))..)..---..... ( -38.40) >DroPer_CAF1 186781 113 + 1 --CUGCGGGGAUUAUGAUAAUAUAGCUCAAGUGCAGUUAGUUGAUGUCGUUGGCGUU---GCCUUUGCUGUUCCCGUUGCAGUUUGCGAUAAUCAUAAUAAUAAUAUCUGUGCUGCUC-- --..(((((((.(((....)))((((..(((.((((...((..((...))..)).))---))))).))))))))))).(((((..((((((.............)))).)))))))..-- ( -26.52) >consensus UUCUGCUAGGAUUAUGAUAAUAUAGCUCAAGUGCAGUUAGUCGAUGUCGCUGCU______GC_GCUGCAGUUGCACUUGCAGUUUGCGAUAAUCAUAAUAAUAAUGGUG______CAA__ ....((((..((((((((......((......))..........((((((..............((((((......))))))...)))))))))))))).....))))............ (-19.74 = -19.83 + 0.09)
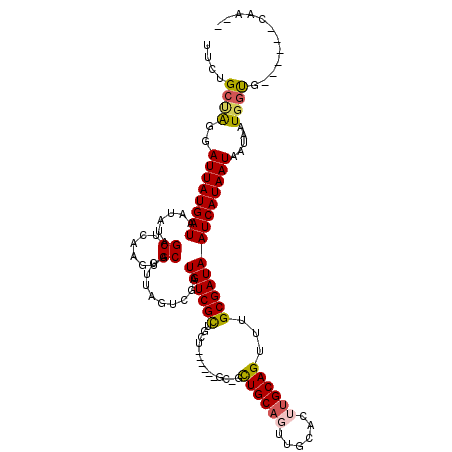
| Location | 12,296,680 – 12,296,776 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 72.50 |
| Mean single sequence MFE | -26.12 |
| Consensus MFE | -10.32 |
| Energy contribution | -10.46 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12296680 96 + 22407834 UCGAUGUCGCUGCUACUGCUGCUGCUGCAGUUGCACUUGCAGUUUGCGAUAAUCAUAAUAAUAAUGGUG------CAA---GUGCUCCGCCGCCGC--C------CCAUCUCC ..((((..((.((....(((((....))))).((((((((.(....)....(((((.......))))))------)))---))))......)).))--.------.))))... ( -30.60) >DroSim_CAF1 123156 90 + 1 UCGAUGUCGCUGCUGCU---GCUGCUGCAUUUGCACUUGCAGUUUGCGAUAAUCAUAAUAAUAAUGGUG------CAA---GUGCUCCUC---CGC--C------CCAUCUCC ..((((..(((((.((.---...)).)))...((((((((.(....)....(((((.......))))))------)))---)))).....---.))--.------.))))... ( -26.40) >DroEre_CAF1 117536 81 + 1 UCGAUGUCGCUG------------CUGCAGUUGCACUUGCAGUUUGCGAUAAUCAUAAUAAUAAUGGUG------CAA---GUGCUCCUC---CGC--C------CCAUCUCC ..((((..((.(------------..(((.((((((((((.....))))....(((.......))))))------)))---.)))..)..---.))--.------.))))... ( -22.80) >DroYak_CAF1 123828 81 + 1 UCGAUGUCGCUG------------CUGCAGUUGCACUUGCAGUUUGCGAUAAUCAUAAUAAUAAUGGUG------CAA---GUGCUCCUC---CGC--C------CCAUCUCC ..((((..((.(------------..(((.((((((((((.....))))....(((.......))))))------)))---.)))..)..---.))--.------.))))... ( -22.80) >DroAna_CAF1 108188 101 + 1 UCGAUGUCGCUGCU------GCAGUUGCACUUGCUCCUGCAGUUUGCGAUAAUCAUAAUAAUAAUGGUGGUG---CUGGAGCCGCUCCUC---CCACUCCCACUCCCACUCCC ..((((((((.(((------((((..((....))..)))))))..))))).)))..........(((..(((---..((((...))))..---.)))..)))........... ( -36.20) >DroPer_CAF1 186819 93 + 1 UUGAUGUCGUUGGCGUU---GCCUUUGCUGUUCCCGUUGCAGUUUGCGAUAAUCAUAAUAAUAAUAUCUGUGCUGCUC---GUGGCCCUG---GGA--U------G---CUUC ..(((((.....)))))---......((...(((((((((.....))))).....................((((...---.))))...)---)))--.------)---)... ( -17.90) >consensus UCGAUGUCGCUGCU______GC_GCUGCAGUUGCACUUGCAGUUUGCGAUAAUCAUAAUAAUAAUGGUG______CAA___GUGCUCCUC___CGC__C______CCAUCUCC ..((((((((..............((((((......))))))...))))).)))........................................................... (-10.32 = -10.46 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:26 2006