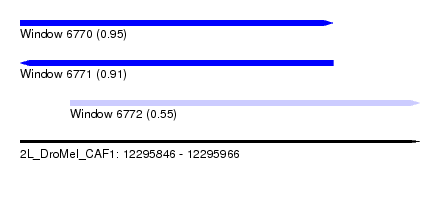
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,295,846 – 12,295,966 |
| Length | 120 |
| Max. P | 0.946255 |

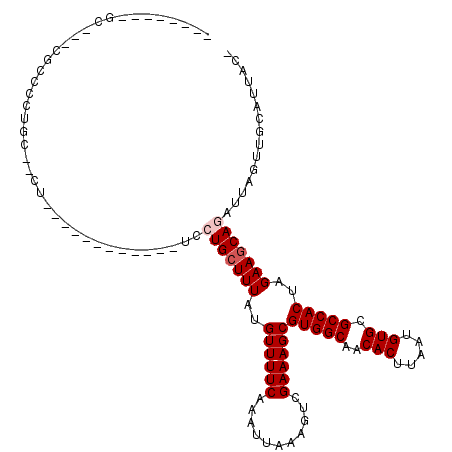
| Location | 12,295,846 – 12,295,940 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.75 |
| Mean single sequence MFE | -29.37 |
| Consensus MFE | -21.15 |
| Energy contribution | -21.65 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946255 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12295846 94 + 22407834 --------GC---CGCCCCUGC--UU------------UUCUGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUUGCAUUAC- --------..---.((....((--(.------------.((((((((..((((((...........))))))(((((..(((.....))).)))))..))))))))..))).)).....- ( -30.90) >DroVir_CAF1 143528 100 + 1 --------GUUGCUGCACCUGCAGCG------------UCAUGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUUGCAUUAAG --------...(((((....)))))(------------((.((((((..((((((...........))))))(((((..(((.....))).)))))..)))))))))............. ( -31.40) >DroWil_CAF1 139660 114 + 1 -CUUUUAUUU---CACCCUUCC--UUUCGCGAUGCUGCUGCUGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUUGCAUUACU -.........---.........--....(..((((.(((((((((((..((((((...........))))))(((((..(((.....))).)))))..)))))))..)))).))))..). ( -32.80) >DroMoj_CAF1 152266 99 + 1 --------GCAGGUGCAGUUGCAGCG------------UCAUGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUUGCAUUA-G --------((....))...(((((((------------((.((((((..((((((...........))))))(((((..(((.....))).)))))..)))))))))..))))))...-. ( -31.60) >DroAna_CAF1 107594 93 + 1 A-------AC---CACCCCUGC--C--------------UCUGGUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUUGCAUUAC- .-------..---.....((((--.--------------((((((..((((((((...........))))))))(((..(((.....))).))))))))).))))..............- ( -25.20) >DroPer_CAF1 185551 91 + 1 ---------------ACCCUCU--UU------------GGCUGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUUGCAUUACC ---------------..((...--..------------))(((((((..((((((...........))))))(((((..(((.....))).)))))..)))))))............... ( -24.30) >consensus ________GC___CGCCCCUGC__CU____________UCCUGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUUGCAUUAC_ ........................................(((((((..((((((...........))))))(((((..(((.....))).)))))..)))))))............... (-21.15 = -21.65 + 0.50)

| Location | 12,295,846 – 12,295,940 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.75 |
| Mean single sequence MFE | -27.58 |
| Consensus MFE | -19.76 |
| Energy contribution | -20.48 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12295846 94 - 22407834 -GUAAUGCAACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAAGCAGAA------------AA--GCAGGGGCG---GC-------- -..............((((((((.(((((((((.....))).))))))((((((..(((((...........)))))..).)------------))--)))))))))---).-------- ( -29.90) >DroVir_CAF1 143528 100 - 1 CUUAAUGCAACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAAGCAUGA------------CGCUGCAGGUGCAGCAAC-------- ..............(((((((...(((((((((.....))).))))))(.((((((.......)))))).)...))))).))------------.(((((....)))))...-------- ( -29.30) >DroWil_CAF1 139660 114 - 1 AGUAAUGCAACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAAGCAGCAGCAGCAUCGCGAAA--GGAAGGGUG---AAAUAAAAG- .(..((((..((...((((((...(((((((((.....))).))))))(.((((((.......)))))).)...)))))).))..))))..)....--.........---.........- ( -28.80) >DroMoj_CAF1 152266 99 - 1 C-UAAUGCAACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAAGCAUGA------------CGCUGCAACUGCACCUGC-------- .-...((((.(...(((((((...(((((((((.....))).))))))(.((((((.......)))))).)...))))).))------------.).))))...........-------- ( -23.50) >DroAna_CAF1 107594 93 - 1 -GUAAUGCAACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAACCAGA--------------G--GCAGGGGUG---GU-------U -.......(((((.(((((((((.(((((((((.....))).))))))(.((((((.......)))))).).......)))--------------)--)))))..))---))-------) ( -31.60) >DroPer_CAF1 185551 91 - 1 GGUAAUGCAACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAAGCAGCC------------AA--AGAGGGU--------------- ...............((((((...(((((((((.....))).))))))(.((((((.......)))))).)...))))))((------------..--...))..--------------- ( -22.40) >consensus _GUAAUGCAACUAAUCUGCUUCUAGUGGCGCACAUUAAGUGUUGCCACGCUUUCGACUUUAAUUUGAAAACAUAAAGCAGAA____________AA__GCAGGGGCG___GC________ ..............(((((((...(((((((((.....))).))))))(.((((((.......)))))).)...)))))))....................................... (-19.76 = -20.48 + 0.72)

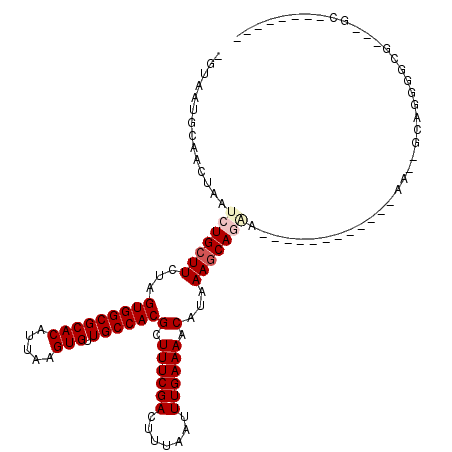
| Location | 12,295,861 – 12,295,966 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.08 |
| Mean single sequence MFE | -26.15 |
| Consensus MFE | -21.95 |
| Energy contribution | -22.15 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.551523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12295861 105 + 22407834 CUGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUUGCAUUAC--------C---C--CGAAG-CCG-GCUCACUGACUCGUCC .((((((..((((((...........))))))(((((..(((.....))).)))))..))))))(((.((((.......--------.---(--((...-.))-)......)))).))). ( -26.64) >DroVir_CAF1 143548 91 + 1 AUGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUUGCAUUAAGGGUGUU-C---C-------------------------CGA .((((((..((((((...........))))))(((((..(((.....))).)))))..))))))...............(((....-)---)-------------------------).. ( -25.50) >DroPse_CAF1 179968 109 + 1 CUGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUUGCAUUACC---UUC-C---C--CGAAG-CCG-GCUCAGUGACUCCUCC (((((((..((((((...........))))))(((((..(((.....))).)))))..)))))))...(((..(......---...-.---(--((...-.))-)....)..)))..... ( -27.44) >DroGri_CAF1 150346 118 + 1 AUGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAAUACUUAAUGUGCGCCACUAGAAGCAGAUUAGUUGCAUUAAGGUUGGC-CUGCCCACGACG-ACGAGCACACACACACAGUA .(((((..((....)).......((((....(((((((((...((((.(((..........))).)))))))))....(((....)-))...)))).))-)))))))............. ( -26.20) >DroWil_CAF1 139694 111 + 1 CUGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUUGCAUUACU---CUACC---C--CUAUCCCCC-GCCCGAUUCCAAGUCC (((((((..((((((...........))))))(((((..(((.....))).)))))..)))))))...............---.....---.--.........-................ ( -23.70) >DroPer_CAF1 185562 109 + 1 CUGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUUGCAUUACC---UUC-C---C--CGAAG-CCG-GCUCAGUGACUCCUCC (((((((..((((((...........))))))(((((..(((.....))).)))))..)))))))...(((..(......---...-.---(--((...-.))-)....)..)))..... ( -27.44) >consensus CUGCUUUAUGUUUUCAAAUUAAAGUCGAAAGCGUGGCAACACUUAAUGUGCGCCACUAGAAGCAGAUUAGUUGCAUUACC___GUC_C___C__CGAAG_CCG_GCUCACUGACUCCUCC (((((((..((((((...........))))))(((((..(((.....))).)))))..)))))))....................................................... (-21.95 = -22.15 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:24 2006