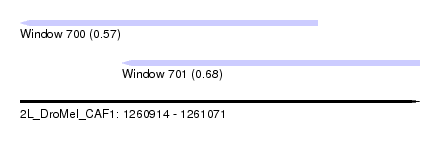
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,260,914 – 1,261,071 |
| Length | 157 |
| Max. P | 0.682625 |

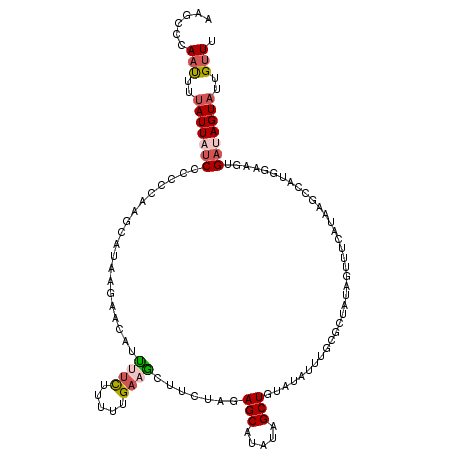
| Location | 1,260,914 – 1,261,031 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.02 |
| Mean single sequence MFE | -28.80 |
| Consensus MFE | -15.32 |
| Energy contribution | -17.12 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1260914 117 - 22407834 CUUUUUGAAGCUUUUAGAGCAUAUAGCUGUAUAUUUGCUCUAUAGUUUCAUAAGCCAUGGAAGUGAUAGUAUUGUUUUCGUGGCUUUGGUAUGAUAACCAAUCCCACAACACGCUUU .....(((((((..(((((((.(((......))).))))))).))))))).((((((((((((..((...))..))))))))))))((((......))))................. ( -37.50) >DroSec_CAF1 20537 117 - 1 UUUUUUGACGCUUCUAGAGCAUAUAGCUGUAUAUUUGCGCUAUAGUUUCAUAAGCCAUGGAAGUGAUAGUAUUGUUUUCGUGGCUUUGGCAUGAUAACCAAUCCCACAACACGCUUU ...............(((((.((((((.(((....)))))))))(((((((((((((((((((..((...))..))))))))))))....)))).)))..............))))) ( -30.40) >DroSim_CAF1 21050 117 - 1 CUUUUUGACGCUUCUAGAGCAUAUAGCUGUAUAUUUGCGCUAUAGUUUCAUAAGCCAUGGAAGUGAUAGUAUUGUUUUCGUGGCUUUGGCAUGAUAACCAAUCCCACAACACGCUUU ...............(((((.((((((.(((....)))))))))(((((((((((((((((((..((...))..))))))))))))....)))).)))..............))))) ( -30.40) >DroEre_CAF1 33144 105 - 1 CCA-CUGAAUCCUCUCGAGCAUACAGCUCCACAUUGGACCUAUAGUUCCA----------UAGUGAUAGUAUUGUUUUGGUGGCUUUGGC-UGAUAACCAAUCCCACAACACGCUUU ..(-(((.........((((.....))))(((..((((.(....).))))----------..))).)))).........((((..((((.-......))))..)))).......... ( -23.30) >DroYak_CAF1 21362 106 - 1 CCUUAUGAAUGUUUUAUAGCAUAUUGCUGU---------GUAUAGUUUAA--AGACAAGCAAGUGAUAGUAUUGUUUUCGUGGCUUUGGCAUGAUAACCAAUCCCACAACGCGCUUU .(((.(((((....((((((.....)))))---------)....))))))--))..(((((((..((...))..)))..((((..((((........))))..)))).....)))). ( -22.40) >consensus CUUUUUGAAGCUUCUAGAGCAUAUAGCUGUAUAUUUGCGCUAUAGUUUCAUAAGCCAUGGAAGUGAUAGUAUUGUUUUCGUGGCUUUGGCAUGAUAACCAAUCCCACAACACGCUUU ................((((....(((((((.........)))))))...........(((((..((...))..)))))((((..((((........))))..)))).....)))). (-15.32 = -17.12 + 1.80)

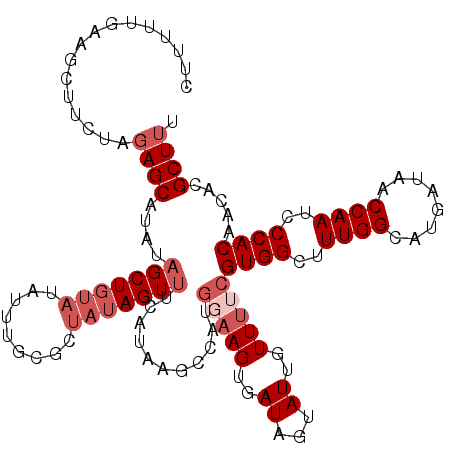
| Location | 1,260,954 – 1,261,071 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 74.49 |
| Mean single sequence MFE | -22.94 |
| Consensus MFE | -3.24 |
| Energy contribution | -4.00 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.14 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1260954 117 - 22407834 AAUCCCAAUUCUAUUAUCCCCACUAGCAUAAGAACAUUUUCUUUUUGAAGCUUUUAGAGCAUAUAGCUGUAUAUUUGCUCUAUAGUUUCAUAAGCCAUGGAAGUGAUAGUAUUGUUU .....((((.(((((((..(((...((..(((((....)))))..(((((((..(((((((.(((......))).))))))).)))))))...))..)))..))))))).))))... ( -32.70) >DroSec_CAF1 20577 117 - 1 AAGCCCACUUUUAUUAUCCCCUCAAGCAUAAGAACAUUUAUUUUUUGACGCUUCUAGAGCAUAUAGCUGUAUAUUUGCGCUAUAGUUUCAUAAGCCAUGGAAGUGAUAGUAUUGUUU .....(((((((((.......(((((.(((((....)))))..))))).((((.(..(((.((((((.(((....))))))))))))..).)))).)))))))))............ ( -22.40) >DroSim_CAF1 21090 117 - 1 AAGCCCAAUUUUAUUAUCCCCCCAAGCAUAAGAACAUUUUCUUUUUGACGCUUCUAGAGCAUAUAGCUGUAUAUUUGCGCUAUAGUUUCAUAAGCCAUGGAAGUGAUAGUAUUGUUU ......(((..((((((((..(((.(((.(((((....)))))..)).)((((.(..(((.((((((.(((....))))))))))))..).))))..)))..).)))))))..))). ( -25.20) >DroEre_CAF1 33183 106 - 1 AAGUUAAACUUCAUUCUCUCCCCAAGUUUAAGCAAUUGUUCCA-CUGAAUCCUCUCGAGCAUACAGCUCCACAUUGGACCUAUAGUUCCA----------UAGUGAUAGUAUUGUUU ...((((((((............))))))))(((((.....((-(((.........((((.....)))).....((((.(....).))))----------))))).....))))).. ( -18.50) >DroYak_CAF1 21402 106 - 1 AAGCCAAAUUUUAUUCCCUCCUCGAGUUUAAGCAAAUAUUCCUUAUGAAUGUUUUAUAGCAUAUUGCUGU---------GUAUAGUUUAA--AGACAAGCAAGUGAUAGUAUUGUUU ....(((...((((..(...((.(..(((((((((((((((.....))))))))((((((.....)))))---------)....))))))--)..).))...)..))))..)))... ( -15.90) >consensus AAGCCCAAUUUUAUUAUCCCCCCAAGCAUAAGAACAUUUUCUUUUUGAAGCUUCUAGAGCAUAUAGCUGUAUAUUUGCGCUAUAGUUUCAUAAGCCAUGGAAGUGAUAGUAUUGUUU ......(((..(((((((...................((((.....)))).......(((.....)))....................................)))))))..))). ( -3.24 = -4.00 + 0.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:04 2006