
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,266,512 – 12,266,618 |
| Length | 106 |
| Max. P | 0.798362 |

| Location | 12,266,512 – 12,266,618 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.55 |
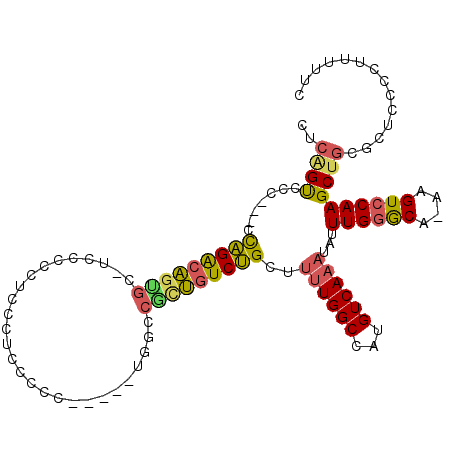
| Mean single sequence MFE | -22.42 |
| Consensus MFE | -18.43 |
| Energy contribution | -18.30 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
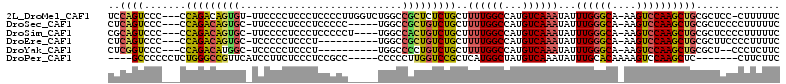
>2L_DroMel_CAF1 12266512 106 + 22407834 UCCAGUCCC---CCAGACAGUGU-UUCCCCUCCCUCCCCUUGGUCUGGCCGCUCUCUGCUUUUGGCCAUGUCAAAUAUUUGGGCA-AAGUCCAAGCUGCGCUCC-CUUUUUC ....(((..---...)))(((((-...............((((..(((((((.....))....)))))..))))...(((((((.-..)))))))..)))))..-....... ( -23.20) >DroSec_CAF1 89665 102 + 1 CUCAGUCCC---CCAGACAGUGC-UUCCCCUCCCUCCCCC-----UGGCCGCUGUCUGCUUUUGGCCAUGUCAAAUAUUUGGGCA-AAGUCCAAGCUGCGCUCCCCUUUUUC ..((((...---.((((((((((-(...............-----.)).)))))))))..((((((...))))))...((((((.-..)))))))))).............. ( -22.79) >DroSim_CAF1 93773 103 + 1 CGCAGUCCC---CCAGACAGUGC-UUCCCCUCCCUCCCCCU----UGGCCACUGUCUGCUUUUGGCCAUGUCAAAUAUUUGGGCA-AAGUCCAAGCUGCGCUCCCCUUUUUC ((((((...---.((((((((((-(................----.)).)))))))))..((((((...))))))...((((((.-..))))))))))))............ ( -28.83) >DroEre_CAF1 87549 97 + 1 CUCAGUCCC---CCAGACAGUGC-UCCCCCUCCCU----------UGGCCGCUGUCUGCUUUUGGCCAUGUCAAAUAUUUGGGCA-AAGUCCAAGCUGCGCUUCCCCUUUUC ..((((...---.(((((((((.-....((.....----------.)).)))))))))..((((((...))))))...((((((.-..)))))))))).............. ( -23.20) >DroYak_CAF1 93392 95 + 1 CUCGGUCCC---CCAGACAUGGC-UCCCCCUCCCU----------UGGCCCCUGUCUGCUUUUGGCCAUGUCAAAUAUUUGGGCA-AAGUCCAAGCUGCGCU--CCCUCUUC ...((((..---.((((((.(((-(..........----------.))))..)))))).....)))).............((((.-.(((....)))..)))--)....... ( -21.80) >DroPer_CAF1 146940 96 + 1 ----GCCCCCCUCUGGGCCGUUCAUCCUUCUCCCUCCGCC-----CCCCCUUGGUCCGCUCAUGGCUAUGUCAAAUAUUUGCACAAAAGUCCAAGCUC-------CUUCUUC ----((((......))))......................-----....(((((.(.((.....))..((((((....))).)))...).)))))...-------....... ( -14.70) >consensus CUCAGUCCC___CCAGACAGUGC_UCCCCCUCCCUCCCCC_____UGGCCGCUGUCUGCUUUUGGCCAUGUCAAAUAUUUGGGCA_AAGUCCAAGCUGCGCUCCCCUUUUUC ..((((.......(((((((((...........................)))))))))..((((((...))))))...((((((....)))))))))).............. (-18.43 = -18.30 + -0.13)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:13 2006