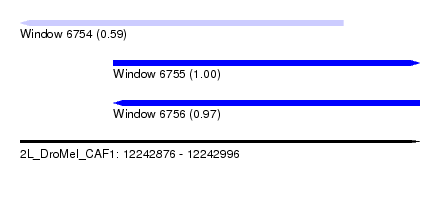
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,242,876 – 12,242,996 |
| Length | 120 |
| Max. P | 0.997960 |

| Location | 12,242,876 – 12,242,973 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 84.68 |
| Mean single sequence MFE | -28.04 |
| Consensus MFE | -16.29 |
| Energy contribution | -18.05 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12242876 97 - 22407834 GUGAGUA-----AAGGAAAGAGUACGGUUUCGUGUCGAGCAUGUGCUCGGAAAACUCGAUUUUCCUCACACCUACGACAACAAACUCACUUAAGUUAACCUA ((((((.-----.(((...(((((((..((((...))))..)))))))((((((.....)))))).....)))..(....)..))))))............. ( -23.90) >DroSec_CAF1 66227 102 - 1 GUGAGGAAGGGAAAGGAAAGAGCACGGUUUCGUGUCGAGCAUGUGCUCGGAAAACUCGAUUUUCCUCACACCUACACCUUCGAACUCACUUAAGUUAACCUA ((((((((((...(((...(((((((..((((...))))..)))))))((((((.....)))))).....)))...)))))...)))))............. ( -33.70) >DroSim_CAF1 67669 102 - 1 GUGAGAAAAGGAAAGGAAAGAGCACGGUUUCGUGUCGAGCAUGUGCUCGGAAAACUCGAUUUUCCUCACACCUACACCUUCGAACUCACUUAAGUUAACCUA (((((..((((..(((...(((((((..((((...))))..)))))))((((((.....)))))).....)))...))))....)))))............. ( -29.40) >DroEre_CAF1 64118 99 - 1 GUACGG--GUGUAAGGAAGGAGU-AGGUUUCGUGUCGAGCAUGUGCUCGGAAAACUCGAUUUUCCUUACACCUACACCUCCGAACACACUUAACUUAACCUA ......--((((..(((.((.((-((((........((((....))))((((((.....))))))....)))))).)))))..))))............... ( -31.00) >DroYak_CAF1 65270 90 - 1 G----------UAAGGAA-AAGC-CGUUUUCGUGUCGAGCAUGUGCUCGGAAAACUCGAUUUUCCUUAUACCUACACCUCCGAACCCACUUAACUUAACGUA (----------(((((((-((..-.((((((....(((((....)))))))))))....))))))))))................................. ( -22.20) >consensus GUGAGGA___GAAAGGAAAGAGCACGGUUUCGUGUCGAGCAUGUGCUCGGAAAACUCGAUUUUCCUCACACCUACACCUCCGAACUCACUUAAGUUAACCUA (((((........(((...(((((((..((((...))))..)))))))((((((.....)))))).....)))...........)))))............. (-16.29 = -18.05 + 1.76)

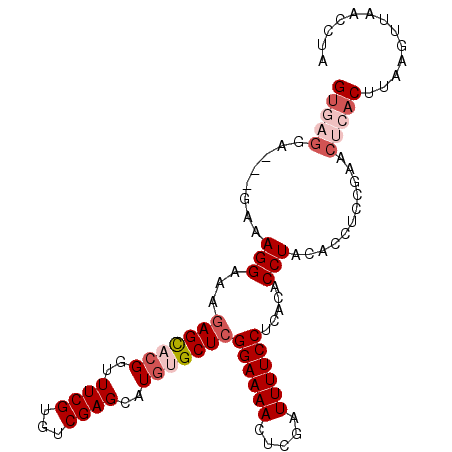
| Location | 12,242,904 – 12,242,996 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 85.26 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -20.10 |
| Energy contribution | -20.46 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -4.64 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12242904 92 + 22407834 UAGGUGUGAGGAAAAUCGAGUUUUCCGAGCACAUGCUCGACACGAAACCGUACUCUUUCCUU-----UACUCACACCCAAACACACCCCCACAGACA ..((((((((((((...((((....(((((....)))))..(((....))))))))))))..-----...))))))).................... ( -25.40) >DroSec_CAF1 66255 95 + 1 UAGGUGUGAGGAAAAUCGAGUUUUCCGAGCACAUGCUCGACACGAAACCGUGCUCUUUCCUUUCCCUUCCUCACACCCAAAU--AUCCCCGCAGACA ..(((((((((((....(((.....(((((....))))).((((....)))).......)))....))))))))))).....--............. ( -31.40) >DroSim_CAF1 67697 95 + 1 UAGGUGUGAGGAAAAUCGAGUUUUCCGAGCACAUGCUCGACACGAAACCGUGCUCUUUCCUUUCCUUUUCUCACACCCAAAC--AUCCCCGCAGACA ..((((((((((((...(((.....(((((....))))).((((....)))).......)))...)))))))))))).....--............. ( -29.70) >DroEre_CAF1 64146 92 + 1 UAGGUGUAAGGAAAAUCGAGUUUUCCGAGCACAUGCUCGACACGAAACCU-ACUCCUUCCUUACAC--CCGUACACCCAAAC--AUCCCCGCAAACA ..(((((((((((....(((((((((((((....)))))....))))...-)))).))))))))))--).............--............. ( -27.30) >DroYak_CAF1 65298 83 + 1 UAGGUAUAAGGAAAAUCGAGUUUUCCGAGCACAUGCUCGACACGAAAACG-GCUU-UUCCUUA----------CACCCAAAC--ACCCCCGCAGACA ..(((.(((((((((.(..(((((((((((....)))))....)))))).-).))-)))))))----------.))).....--............. ( -25.80) >consensus UAGGUGUGAGGAAAAUCGAGUUUUCCGAGCACAUGCUCGACACGAAACCGUGCUCUUUCCUUAC___UCCUCACACCCAAAC__AUCCCCGCAGACA ..((((((((((((...(((((((((((((....)))))....))))....))))))))))..........)))))).................... (-20.10 = -20.46 + 0.36)

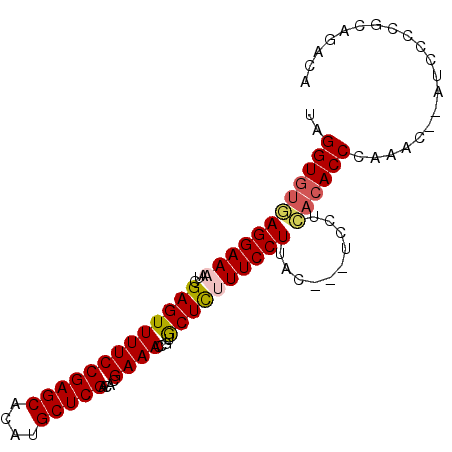
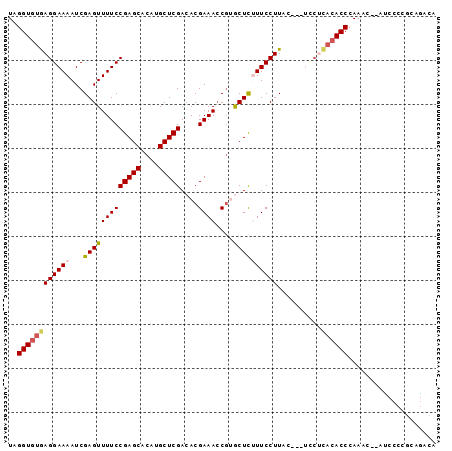
| Location | 12,242,904 – 12,242,996 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 85.26 |
| Mean single sequence MFE | -32.72 |
| Consensus MFE | -23.24 |
| Energy contribution | -24.20 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
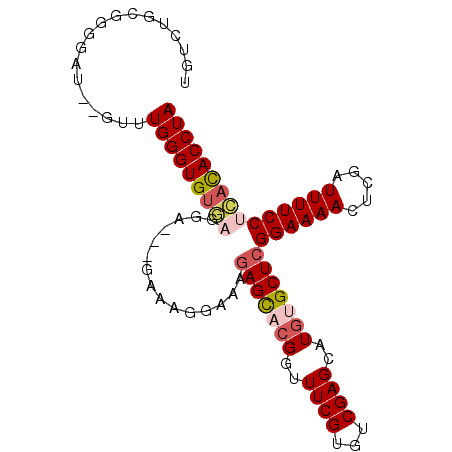
>2L_DroMel_CAF1 12242904 92 - 22407834 UGUCUGUGGGGGUGUGUUUGGGUGUGAGUA-----AAGGAAAGAGUACGGUUUCGUGUCGAGCAUGUGCUCGGAAAACUCGAUUUUCCUCACACCUA ..................((((((((((.(-----(((....(((((((....))).((((((....))))))...))))..)))).)))))))))) ( -30.70) >DroSec_CAF1 66255 95 - 1 UGUCUGCGGGGAU--AUUUGGGUGUGAGGAAGGGAAAGGAAAGAGCACGGUUUCGUGUCGAGCAUGUGCUCGGAAAACUCGAUUUUCCUCACACCUA (((((....))))--)..(((((((((((((((...((....(((((((..((((...))))..)))))))......))...))))))))))))))) ( -38.20) >DroSim_CAF1 67697 95 - 1 UGUCUGCGGGGAU--GUUUGGGUGUGAGAAAAGGAAAGGAAAGAGCACGGUUUCGUGUCGAGCAUGUGCUCGGAAAACUCGAUUUUCCUCACACCUA .((((....))))--...(((((((((.....((((((....(((((((....))))((((((....))))))....)))..))))))))))))))) ( -32.60) >DroEre_CAF1 64146 92 - 1 UGUUUGCGGGGAU--GUUUGGGUGUACGG--GUGUAAGGAAGGAGU-AGGUUUCGUGUCGAGCAUGUGCUCGGAAAACUCGAUUUUCCUUACACCUA .............--............((--((((((((((((...-.(((((....((((((....)))))).)))))...)))))))))))))). ( -30.80) >DroYak_CAF1 65298 83 - 1 UGUCUGCGGGGGU--GUUUGGGUG----------UAAGGAA-AAGC-CGUUUUCGUGUCGAGCAUGUGCUCGGAAAACUCGAUUUUCCUUAUACCUA .............--...((((((----------(((((((-((..-.((((((....(((((....)))))))))))....))))))))))))))) ( -31.30) >consensus UGUCUGCGGGGAU__GUUUGGGUGUGAGGA___GAAAGGAAAGAGCACGGUUUCGUGUCGAGCAUGUGCUCGGAAAACUCGAUUUUCCUCACACCUA ..................(((((((((...............(((((((..((((...))))..)))))))((((((.....))))))))))))))) (-23.24 = -24.20 + 0.96)

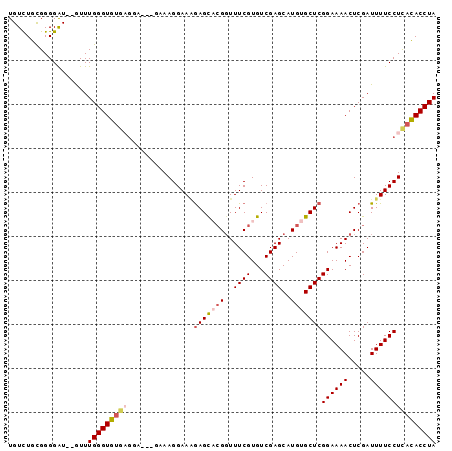
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:09 2006