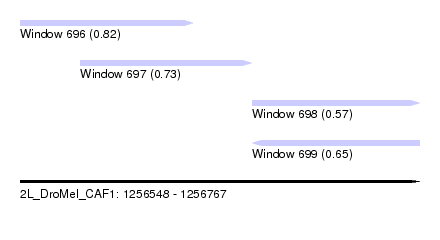
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,256,548 – 1,256,767 |
| Length | 219 |
| Max. P | 0.816687 |

| Location | 1,256,548 – 1,256,643 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 85.88 |
| Mean single sequence MFE | -29.93 |
| Consensus MFE | -25.83 |
| Energy contribution | -25.17 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
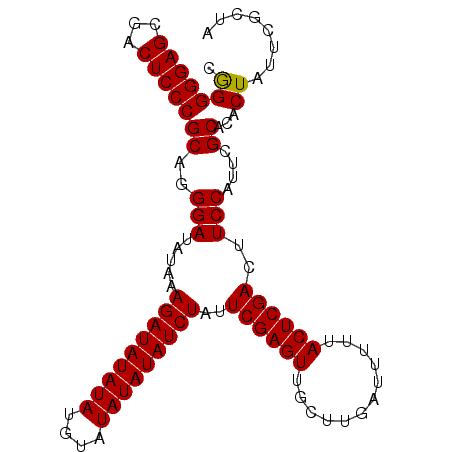
>2L_DroMel_CAF1 1256548 95 + 22407834 AAAUC--AGCUAAUUGACACAACGGACAUGCGAGGGCCAGACUUUGGCCAAAU----UUGGCCUGGCCAAAAAGGAAGCGGAAGUACUGCGGCCGCAAAAU .....--.....................(((...(((((......(((((...----.)))))))))).....((..((((.....))))..))))).... ( -27.30) >DroSec_CAF1 16240 94 + 1 AAAUC--AGCUAAUUGACACUACGGACAUGCGACGGCCAGACUUUGGCCAAAU----UUGGCCUGGCCAA-AAGGAAGCGGAAGCACUGCGUCCGCAAAAU .....--...............(((((..(((..(((((......(((((...----.))))))))))..-......((....))..))))))))...... ( -29.90) >DroSim_CAF1 16732 94 + 1 AAAUC--AGCUAAUUGACACUACGGACAUGCGACGGCCAGACUUUGGCCAAAU----UUGGCCUGGCCAA-AAGGAAGCGGAAGCACUGCGGCCGCAAAAU .....--...............(((...((((..(((((......(((((...----.))))))))))..-......((....))..)))).)))...... ( -28.30) >DroEre_CAF1 28965 94 + 1 AAAUC--AGCUAAUUGACACUGCGGACAUGCGACGGCCAGAUUUUGGCCAAAU----UUGGCCUGGCCAA-AAGGAAGCGGAAGCACUGUGGCCGCAACAU ...((--.((...........)).))..(((...((((((((((.....))))----)))))).(((((.-..((..((....)).)).)))))))).... ( -32.20) >DroYak_CAF1 16971 96 + 1 AAAUCUAAGCUAAUUGACAUUACGGACAUGCGAGGGCCGGACUUUGGCCAAAU----UUGGCCUGGCCAA-AAGGAAGCGGAAGCAUUGUGGCCGCAACAU ........((...(((.(((.......)))))).((((((.((..(((((...----.))))).))))..-......((....)).....))))))..... ( -28.00) >DroAna_CAF1 40041 97 + 1 AGGCC--UGCUAAUUGAUA-UACGAACAUGUGGCAGCUAGACUCUGGCCAGAAGUUCCUGGCCUGGCCGA-AAGGAAGCGGAAGCGUUUAAGCCGCACAAU .((((--(((((..((...-......))..)))))).(((((...((((((......))))))...((..-..))..((....))))))).)))....... ( -33.90) >consensus AAAUC__AGCUAAUUGACACUACGGACAUGCGACGGCCAGACUUUGGCCAAAU____UUGGCCUGGCCAA_AAGGAAGCGGAAGCACUGCGGCCGCAAAAU ............................((((..(((((......((((((......))))))))))).........((....))........)))).... (-25.83 = -25.17 + -0.66)

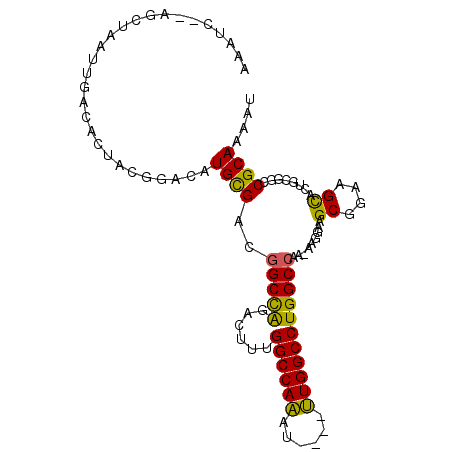
| Location | 1,256,581 – 1,256,675 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 81.22 |
| Mean single sequence MFE | -29.23 |
| Consensus MFE | -21.36 |
| Energy contribution | -21.17 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1256581 94 + 22407834 GCCAGACUUUGGCCAAAU----UUGGCCUGGCCAAAAAGGAAGCGGAAGUACUGCGGCCGCAAAAUACAUUGUAAACU-------GCUAAACUGCAGGGGAAGUG .....(((((((((...(----(((((...))))))......((((.....))))))))((((......))))...((-------((......))))..))))). ( -28.50) >DroSec_CAF1 16273 100 + 1 GCCAGACUUUGGCCAAAU----UUGGCCUGGCCAA-AAGGAAGCGGAAGCACUGCGUCCGCAAAAUACAUUGUAAACUGCUAACUGCUAAACUGCAGGGGAAGUG .....((((((((((...----.)))))((((...-..(((.((((.....)))).)))((((......)))).....)))).((((......))))..))))). ( -29.60) >DroSim_CAF1 16765 100 + 1 GCCAGACUUUGGCCAAAU----UUGGCCUGGCCAA-AAGGAAGCGGAAGCACUGCGGCCGCAAAAUACAUUGUAAACUGCUAACUGCUAAACUGCAGGGGAAGUG .....((((((((((...----.))))).((((..-.((...((....)).))..))))(((...(((...)))...)))...((((......))))..))))). ( -31.00) >DroEre_CAF1 28998 93 + 1 GCCAGAUUUUGGCCAAAU----UUGGCCUGGCCAA-AAGGAAGCGGAAGCACUGUGGCCGCAACAUACAUUGUAAACU-------CCUAAACUGCAGGGGAAGUG ((........(((((...----.))))).(((((.-..((..((....)).)).))))))).(((.....)))...((-------(((.......)))))..... ( -29.60) >DroYak_CAF1 17006 93 + 1 GCCGGACUUUGGCCAAAU----UUGGCCUGGCCAA-AAGGAAGCGGAAGCAUUGUGGCCGCAACAUACAUUGUAAACU-------CCUAAACUGCAGGCGAAGUG (((((.((..(((((...----.))))).))))..-.((((.((((..((...))..)))).(((.....)))....)-------)))........)))...... ( -30.00) >DroAna_CAF1 40073 86 + 1 GCUAGACUCUGGCCAGAAGUUCCUGGCCUGGCCGA-AAGGAAGCGGAAGCGUUUAAGCCGCACAAUGACGUACAAACUG-------C--AACU---------GUG (((((((...((((((......))))))...((..-..))..((....)))))).))).(((...((.....))...))-------)--....---------... ( -26.70) >consensus GCCAGACUUUGGCCAAAU____UUGGCCUGGCCAA_AAGGAAGCGGAAGCACUGCGGCCGCAAAAUACAUUGUAAACU_______GCUAAACUGCAGGGGAAGUG .....(((((((((((......)))))).((((....((...((....)).))..))))((((......))))..........................))))). (-21.36 = -21.17 + -0.19)
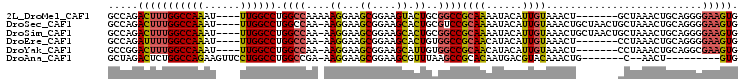
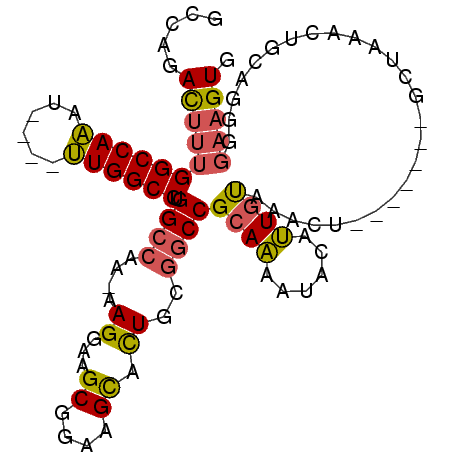
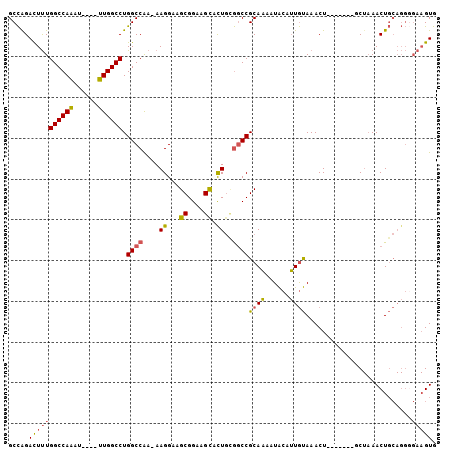
| Location | 1,256,675 – 1,256,767 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -23.33 |
| Consensus MFE | -23.73 |
| Energy contribution | -23.90 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.20 |
| Structure conservation index | 1.02 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1256675 92 + 22407834 UAGCGAAUAGUGUGCGAAUGGAAGUCGAGUAAAAAUCAAGCAACUCGAAUAGAUAUA--------UAUAUCUUUAUAUCCCUGCGGGAGUCGCUCCCCUG .(((((....(.((((...((...((((((............))))))...((((((--------........)))))))))))).)..)))))...... ( -20.40) >DroSec_CAF1 16373 100 + 1 UAGCGAAUAGUGUGCGAAUGGAAGUCGAGUAAAAAUCAAGCAACUCGAAUAGAUAUAUAUACAUAUAUAUCUUUAUAUCCCUGCGGGAGUCGCUCCCCCG .(((((....(.((((...(((..((((((............))))))..(((((((((....))))))))).....))).)))).)..)))))...... ( -24.80) >DroSim_CAF1 16865 100 + 1 UAGCGAAUAGUGUGCGAAUGGAAGUCGAGUAAAAAUCAAGCAACUCGAAUAGAUAUAUAUACAUAUAUAUCUUUAUAUCCCUGCGGGAGUCGCUCCCCCG .(((((....(.((((...(((..((((((............))))))..(((((((((....))))))))).....))).)))).)..)))))...... ( -24.80) >consensus UAGCGAAUAGUGUGCGAAUGGAAGUCGAGUAAAAAUCAAGCAACUCGAAUAGAUAUAUAUACAUAUAUAUCUUUAUAUCCCUGCGGGAGUCGCUCCCCCG .(((((....(.((((...(((..((((((............))))))..(((((((((....))))))))).....))).)))).)..)))))...... (-23.73 = -23.90 + 0.17)

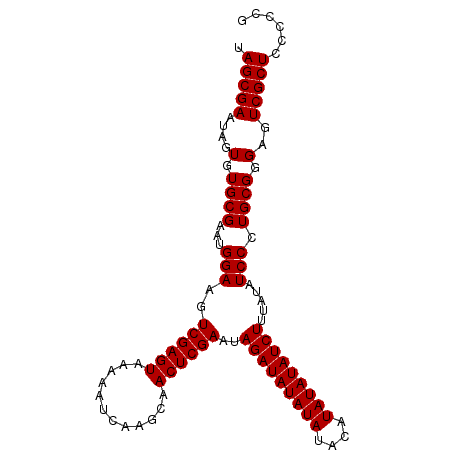
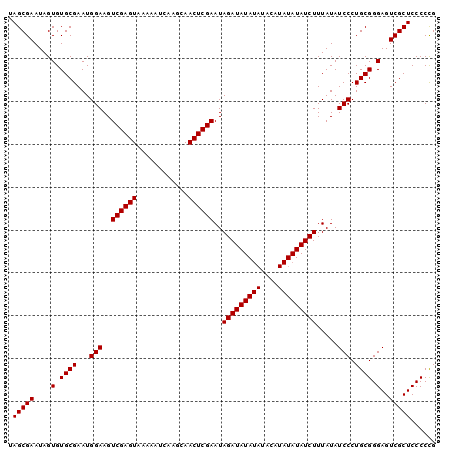
| Location | 1,256,675 – 1,256,767 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -23.65 |
| Energy contribution | -23.60 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.38 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1256675 92 - 22407834 CAGGGGAGCGACUCCCGCAGGGAUAUAAAGAUAUA--------UAUAUCUAUUCGAGUUGCUUGAUUUUUACUCGACUUCCAUUCGCACACUAUUCGCUA ...(((((...)))))((..(((((((........--------)))))))..((((((............)))))).........))............. ( -20.30) >DroSec_CAF1 16373 100 - 1 CGGGGGAGCGACUCCCGCAGGGAUAUAAAGAUAUAUAUGUAUAUAUAUCUAUUCGAGUUGCUUGAUUUUUACUCGACUUCCAUUCGCACACUAUUCGCUA .(.(((((...))))).).((((.....(((((((((....)))))))))..((((((............)))))).))))................... ( -25.40) >DroSim_CAF1 16865 100 - 1 CGGGGGAGCGACUCCCGCAGGGAUAUAAAGAUAUAUAUGUAUAUAUAUCUAUUCGAGUUGCUUGAUUUUUACUCGACUUCCAUUCGCACACUAUUCGCUA .(.(((((...))))).).((((.....(((((((((....)))))))))..((((((............)))))).))))................... ( -25.40) >consensus CGGGGGAGCGACUCCCGCAGGGAUAUAAAGAUAUAUAUGUAUAUAUAUCUAUUCGAGUUGCUUGAUUUUUACUCGACUUCCAUUCGCACACUAUUCGCUA .(((((((...)))))((..(((.....(((((((((....)))))))))..((((((............))))))..)))....))...))........ (-23.65 = -23.60 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:02 2006