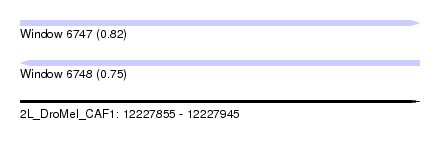
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,227,855 – 12,227,945 |
| Length | 90 |
| Max. P | 0.820465 |

| Location | 12,227,855 – 12,227,945 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 79.65 |
| Mean single sequence MFE | -30.33 |
| Consensus MFE | -15.70 |
| Energy contribution | -16.07 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12227855 90 + 22407834 ------CGCCAGCCAAUGGAAAAGCACAUCGUGUGCUAUGGAAACGGUUUGACCAGCUGGCACAAAUGAAUG--GC-------------GGCAGAGGGAAAAUUGGACUGA ------...((((((((((...(((((.....))))).((....))......)).((((.((........))--.)-------------))).........))))).))). ( -26.30) >DroSec_CAF1 51387 90 + 1 ------UGCCAGCCAAUGGAAAAGCACAUCGUGUGCUAUGGAAACGGUUUGACCAGCUGGCACAAAUGAAUGGGGC-------------GGCU--GGGAAAAUUGGACCGA ------..(((.....)))...(((((.....))))).(((...((((((..(((((((.(.((......)).).)-------------))))--))..))))))..))). ( -31.90) >DroSim_CAF1 52171 88 + 1 ------UGCCAGCCAAUGGAAAAGCACAUCGUGUGCUAUGGAAAUGGUUUGACCAGCUGGCACAAAUGAAUG--GC-------------GGCU--GGGAAAAUUGGACCGA ------..(((.....)))...(((((.....))))).(((...((((((..(((((((.((........))--.)-------------))))--))..))))))..))). ( -29.50) >DroEre_CAF1 49339 88 + 1 ------UGCCAGCCAAUGGAAAAGCACAUUGUGUGCCAUGGAAACGGUUUGACCAGCUGGCACAAAUGAAUG--GC-------------GGCA--GGGAAAAUUGGCCCGA ------((((.((((.((......))((((.(((((((.(....)((.....))...)))))))))))..))--))-------------))))--(((........))).. ( -31.80) >DroYak_CAF1 50302 88 + 1 ------CACCAGCCAAUGGAAAAGCACGUUGUGUGCUAUGCAAACGAUUUGACCAGCUGGCACAAAUGAAUG--GC-------------GGCA--GGGAAAAUCGGACCGA ------..(((.....)))...(((((.....))))).......((((((..((.((((.((........))--.)-------------))).--))..))))))...... ( -26.50) >DroAna_CAF1 47259 107 + 1 UUGUGAUGUCCGCAAACGGAGAAUAAUUUUGUGUACUGUGGCAACUGUUUGGCCAGCUGGCACAAAUGUUUG--GCCAAAAGGCGGGCAGGCG--GGCAAAAAAGGCCUGG ......(((((((....((.(((((..(((((((....((((.........))))....)))))))))))).--.)).....)))))))..((--(((.......))))). ( -36.00) >consensus ______UGCCAGCCAAUGGAAAAGCACAUCGUGUGCUAUGGAAACGGUUUGACCAGCUGGCACAAAUGAAUG__GC_____________GGCA__GGGAAAAUUGGACCGA .......((((((...(((...(((((.....)))))..(....).......)))))))))..................................((..........)).. (-15.70 = -16.07 + 0.36)



| Location | 12,227,855 – 12,227,945 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 79.65 |
| Mean single sequence MFE | -24.34 |
| Consensus MFE | -15.25 |
| Energy contribution | -15.31 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12227855 90 - 22407834 UCAGUCCAAUUUUCCCUCUGCC-------------GC--CAUUCAUUUGUGCCAGCUGGUCAAACCGUUUCCAUAGCACACGAUGUGCUUUUCCAUUGGCUGGCG------ ...................(((-------------((--((....((((.(((....)))))))..........(((((.....))))).......)))).))).------ ( -21.20) >DroSec_CAF1 51387 90 - 1 UCGGUCCAAUUUUCCC--AGCC-------------GCCCCAUUCAUUUGUGCCAGCUGGUCAAACCGUUUCCAUAGCACACGAUGUGCUUUUCCAUUGGCUGGCA------ .((((...........--.)))-------------).............((((((((((.....))........(((((.....)))))........))))))))------ ( -23.30) >DroSim_CAF1 52171 88 - 1 UCGGUCCAAUUUUCCC--AGCC-------------GC--CAUUCAUUUGUGCCAGCUGGUCAAACCAUUUCCAUAGCACACGAUGUGCUUUUCCAUUGGCUGGCA------ ((((.(((((....((--(((.-------------((--((......)).))..)))))...............(((((.....))))).....)))))))))..------ ( -23.40) >DroEre_CAF1 49339 88 - 1 UCGGGCCAAUUUUCCC--UGCC-------------GC--CAUUCAUUUGUGCCAGCUGGUCAAACCGUUUCCAUGGCACACAAUGUGCUUUUCCAUUGGCUGGCA------ ..(((........)))--.(((-------------((--((..(((((((((((..(((...........)))))))))).))))...........)))).))).------ ( -26.62) >DroYak_CAF1 50302 88 - 1 UCGGUCCGAUUUUCCC--UGCC-------------GC--CAUUCAUUUGUGCCAGCUGGUCAAAUCGUUUGCAUAGCACACAACGUGCUUUUCCAUUGGCUGGUG------ ((((((((((((..((--.((.-------------((--((......)).))..)).))..)))))).......(((((.....)))))........))))))..------ ( -23.30) >DroAna_CAF1 47259 107 - 1 CCAGGCCUUUUUUGCC--CGCCUGCCCGCCUUUUGGC--CAAACAUUUGUGCCAGCUGGCCAAACAGUUGCCACAGUACACAAAAUUAUUCUCCGUUUGCGGACAUCACAA .(((((..........--.)))))...(((....)))--.......(((((.((((((......)))))).)))))...............((((....))))........ ( -28.20) >consensus UCGGUCCAAUUUUCCC__UGCC_____________GC__CAUUCAUUUGUGCCAGCUGGUCAAACCGUUUCCAUAGCACACAAUGUGCUUUUCCAUUGGCUGGCA______ .................................................(((((((..((..............(((((.....))))).....))..)))))))...... (-15.25 = -15.31 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:02 2006