
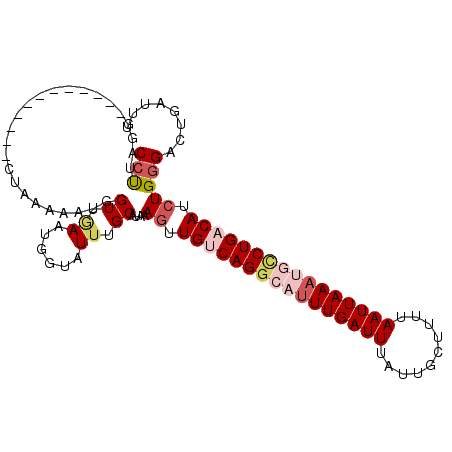
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,224,592 – 12,224,718 |
| Length | 126 |
| Max. P | 0.816015 |

| Location | 12,224,592 – 12,224,687 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 82.56 |
| Mean single sequence MFE | -24.38 |
| Consensus MFE | -15.85 |
| Energy contribution | -17.27 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12224592 95 + 22407834 CCUUAGG------------CUAAAAAUGGUUGAAUGGUAUUUGCAUAAAGUUGUCAGGCAUUUGAUUUAUUGCUUUUAAUUAAAUGCCUGACAUCUGGGACUGAUUU .((((((------------(...(((((.(.....).))))))).......((((((((((((((((..........)))))))))))))))).)))))........ ( -26.30) >DroPse_CAF1 76578 107 + 1 CCUGGGGGGCCUCUGGGCGGCAAAAUUUGUUGAGAAUUAUUUGCAUAAAGUUGUCAGGCAUUUGAUUUAUAGCAUUUAAUUAAAUGCCUGACAUGUGUGACUGGUUU ((..(((...)))..))(((((.....))))).((((((.(..((((....((((((((((((((((..........))))))))))))))))))))..).)))))) ( -36.50) >DroSec_CAF1 48203 95 + 1 CCUUAGG------------CUAAAAAUGGUUGAAUGGUAUUUGCAUAAAGUUGUCAGGCAUUUGAUUUAUUGCUUUUAAUUAAAUGCCUGACAUCUGGGACUGAUUU .((((((------------(...(((((.(.....).))))))).......((((((((((((((((..........)))))))))))))))).)))))........ ( -26.30) >DroSim_CAF1 46973 95 + 1 CCUUAGG------------CUAAAAAUGGUUGAAUGGUAUUUGCAUAAAGUUGUCAGGCAUUUGAUUUAUUGCUUUUAAUUAAAUGUCUGACAUCUGGGACUGAUUU .((((((------------(...(((((.(.....).))))))).......((((((((((((((((..........)))))))))))))))).)))))........ ( -23.60) >DroEre_CAF1 46155 92 + 1 CCUAAGG------------CUACAAAUUGUUCAACGAUAUUUGCAUAAAGUUGUCAGGCAUUUGAUUUAUCGCAUUUAAUUAAAACUCUGUCAUCUGGGGCUGA--- .....((------------(..(..((((.....))))..........((.((.((((..(((((((..........)))))))..)))).)).)))..)))..--- ( -12.70) >DroYak_CAF1 47098 94 + 1 CCCAAGG------------CUAAAAAUUGUUAAACAAUAUUUGCAUAUAGUUGUCAGGCAUUUGAUUUAUUGCUUUUAAUUAA-AUGCUGUCAUCUGGGGCUGAAUU (((...(------------(.(((.((((.....)))).)))))...(((.((.(((.(((((((((..........))))))-)))))).)).))))))....... ( -20.90) >consensus CCUUAGG____________CUAAAAAUGGUUGAAUGGUAUUUGCAUAAAGUUGUCAGGCAUUUGAUUUAUUGCUUUUAAUUAAAUGCCUGACAUCUGGGACUGAUUU (((.........................((.((......)).))....((.((((((((((((((((..........)))))))))))))))).)))))........ (-15.85 = -17.27 + 1.42)


| Location | 12,224,619 – 12,224,718 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.57 |
| Mean single sequence MFE | -26.29 |
| Consensus MFE | -15.80 |
| Energy contribution | -17.13 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.816015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 12224619 99 + 22407834 UUUGCAUAAAGUUGUCAGGCAUUUGAUUUAUUGCUUUUAAUUAAAUGCCUGACAUCUGGGACUGAUUUUAAAUUCUAA-GGUGGAGAUGA-------GUGCGUGAUG .(..(....((.((((((((((((((((..........)))))))))))))))).)).(.(((.(((((.........-....))))).)-------)).))..).. ( -29.92) >DroPse_CAF1 76617 107 + 1 UUUGCAUAAAGUUGUCAGGCAUUUGAUUUAUAGCAUUUAAUUAAAUGCCUGACAUGUGUGACUGGUUUUUGGAUCCAAUGCCAGAGAUGAGAGACGAGUGCGUGAUG ...((((.....((((((((((((((((..........))))))))))))))))..(((..((.((((((((........)))))))).))..))).))))...... ( -37.40) >DroSec_CAF1 48230 99 + 1 UUUGCAUAAAGUUGUCAGGCAUUUGAUUUAUUGCUUUUAAUUAAAUGCCUGACAUCUGGGACUGAUUUUAAAUUCUAA-GGUGGAGAUGA-------GUGCGUGAUU .(..(....((.((((((((((((((((..........)))))))))))))))).)).(.(((.(((((.........-....))))).)-------)).))..).. ( -29.92) >DroSim_CAF1 47000 99 + 1 UUUGCAUAAAGUUGUCAGGCAUUUGAUUUAUUGCUUUUAAUUAAAUGUCUGACAUCUGGGACUGAUUUUAAAUUCUAA-GGUGGAGAUGA-------GUGCGUGAUG .(..(....((.((((((((((((((((..........)))))))))))))))).)).(.(((.(((((.........-....))))).)-------)).))..).. ( -27.22) >DroEre_CAF1 46182 85 + 1 UUUGCAUAAAGUUGUCAGGCAUUUGAUUUAUCGCAUUUAAUUAAAACUCUGUCAUCUGGGGCUGA--------UGGAA-GGUGGAGAUGA-------------GAUG (((.(((....(..((((...(((((((..........))))))).(((........))).))))--------..)..-.))).)))...-------------.... ( -13.40) >DroYak_CAF1 47125 92 + 1 UUUGCAUAUAGUUGUCAGGCAUUUGAUUUAUUGCUUUUAAUUAA-AUGCUGUCAUCUGGGGCUGAAUUUAAACUAUAA-GGUGGAGAUGA-------------AAUG ((..(.(((((((.((((((((((((((..........))))))-)))))(((......)))))).....))))))).-.)..)).....-------------.... ( -19.90) >consensus UUUGCAUAAAGUUGUCAGGCAUUUGAUUUAUUGCUUUUAAUUAAAUGCCUGACAUCUGGGACUGAUUUUAAAUUCUAA_GGUGGAGAUGA_______GUGCGUGAUG .........((.((((((((((((((((..........)))))))))))))))).)).................................................. (-15.80 = -17.13 + 1.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:14:00 2006