
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 12,220,038 – 12,220,141 |
| Length | 103 |
| Max. P | 0.985465 |

| Location | 12,220,038 – 12,220,141 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 58.89 |
| Mean single sequence MFE | -31.91 |
| Consensus MFE | -13.06 |
| Energy contribution | -14.07 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
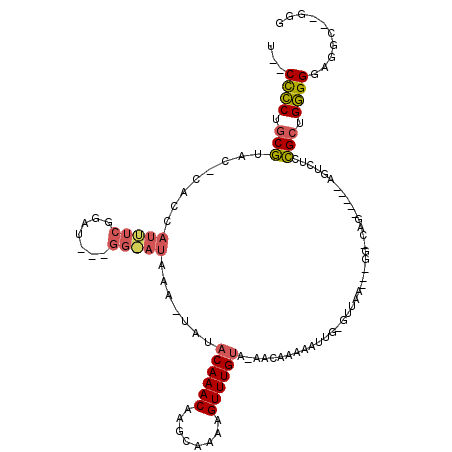
>2L_DroMel_CAF1 12220038 103 + 22407834 U--CCCCUGCGUUC-CCCCAUUUCGGAU---GGAAUAUA-UAUACAAACAAGCAAAAGUUUGUA-AACAAAAAUUGUGUUAAA--GGCCAG-----ACUCUCCGCUGGGGGAGGU--GGG .--..((..(.(((-(((((...(((((---((......-.((((((((((((....)))))).-........))))))....--..))).-----....)))).))))))))).--.)) ( -33.52) >DroPse_CAF1 69241 118 + 1 UAACCCCUUCGUACGCUCCAUCUCUGGUUUGGGGAUAGAGAAUACAAACAAGCAAAAGUUUGCAAAACAAAAAUUCAGUUAAGAGGGGCAGGAGAUAGAGCGCGCUGGGGGGAGG--GGC ...(((((..(..(((((.((((((.(((((.............)))))..((((....))))...........................)))))).)))))..)..)))))...--... ( -36.82) >DroMoj_CAF1 54157 93 + 1 U--CUGCUGCCAGC-AAACGCUUCGCAC---------AA-UACACAAACAAGUAAAAGUUUGUA-AACAAAAAUUG--UUGA------CGG-----GGUCUCGGCUGCGGGUGGCGUAGG .--..((..((.((-(..((((((((..---------..-...((((((........)))))).-((((.....))--))).------)))-----))...))..))).))..))..... ( -25.40) >consensus U__CCCCUGCGUAC_CACCAUUUCGGAU___GG_AUAAA_UAUACAAACAAGCAAAAGUUUGUA_AACAAAAAUUG_GUUAA___GG_CAG_____AGUCUCCGCUGGGGGAGGC__GGG ...((((.(((........(((((.......))))).......((((((........)))))).......................................))).)))).......... (-13.06 = -14.07 + 1.01)

| Location | 12,220,038 – 12,220,141 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 58.89 |
| Mean single sequence MFE | -28.87 |
| Consensus MFE | -14.06 |
| Energy contribution | -13.07 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.49 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
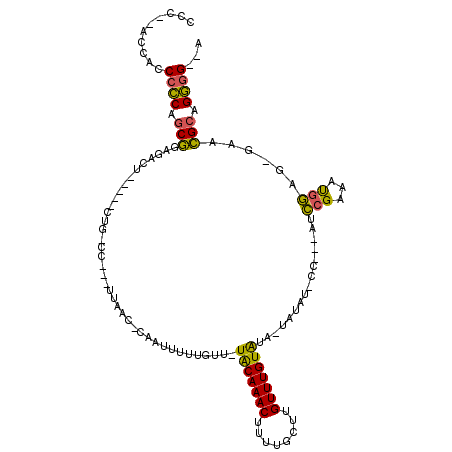
>2L_DroMel_CAF1 12220038 103 - 22407834 CCC--ACCUCCCCCAGCGGAGAGU-----CUGGCC--UUUAACACAAUUUUUGUU-UACAAACUUUUGCUUGUUUGUAUA-UAUAUUCC---AUCCGAAAUGGGG-GAACGCAGGGG--A (((--...(((((((.((((((((-----.((...--.....)).......(((.-(((((((........))))))).)-)).)))).---.))))...)))))-))......)))--. ( -31.30) >DroPse_CAF1 69241 118 - 1 GCC--CCUCCCCCCAGCGCGCUCUAUCUCCUGCCCCUCUUAACUGAAUUUUUGUUUUGCAAACUUUUGCUUGUUUGUAUUCUCUAUCCCCAAACCAGAGAUGGAGCGUACGAAGGGGUUA ...--...((((...(..(((((((((((.((....((......))..........(((((((........)))))))................)))))))))))))..)...))))... ( -35.20) >DroMoj_CAF1 54157 93 - 1 CCUACGCCACCCGCAGCCGAGACC-----CCG------UCAA--CAAUUUUUGUU-UACAAACUUUUACUUGUUUGUGUA-UU---------GUGCGAAGCGUUU-GCUGGCAGCAG--A .....((..((.((((.((.(((.-----..)------))..--....((((((.-(((((((........)))))))..-..---------..)))))))).))-)).))..))..--. ( -20.10) >consensus CCC__ACCACCCCCAGCGGAGACU_____CUG_CC___UUAAC_CAAUUUUUGUU_UACAAACUUUUGCUUGUUUGUAUA_UAUAU_CC___AUCCGAAAUGGAG_GAACGCAGGGG__A ..........((((.(((......................................(((((((........)))))))................(((...)))......))).))))... (-14.06 = -13.07 + -0.99)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:13:57 2006